Association between clinical outcomes and BH3 profile (Individual concentrations)
Junyan Lu
2020-09-04
Last updated: 2020-09-04
Checks: 6 1
Knit directory: BH3profiling/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200826) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 90ada8f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/drugResponse_baseBH3_analysis_AUC_cache/
Ignored: analysis/drugResponse_baseBH3_analysis_cache/
Ignored: analysis/landscape_baseBH3_analysis_AUC_cache/
Ignored: analysis/landscape_baseBH3_analysis_cache/
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/Annexin_rawInput.Rmd
Untracked: analysis/BH3baseline_rawInput.Rmd
Untracked: analysis/BH3dynamic_rawInput.Rmd
Untracked: analysis/based_pep_var.pdf
Untracked: analysis/compareBaseline.Rmd
Untracked: analysis/compareBaseline.pdf
Untracked: analysis/deprecated/
Untracked: analysis/drugResponse_baseBH3_analysis.Rmd
Untracked: analysis/drugResponse_baseBH3_analysis_AUC.Rmd
Untracked: analysis/landscape_baseBH3_analysis.Rmd
Untracked: analysis/landscape_baseBH3_analysis_AUC.Rmd
Untracked: analysis/outcome_baseBH3_analysis.Rmd
Untracked: analysis/patBack.csv
Untracked: analysis/platePlot.pdf
Untracked: analysis/platePlot_dynamic.pdf
Untracked: analysis/predictOutcomes.Rmd
Untracked: code/utils.R
Untracked: data/Data for Thorsten .xlsx
Untracked: data/Raw data Baseline for Junyan.xlsx
Untracked: data/Raw data DBP for Junyan.xlsx
Untracked: data/T. Zenz data for Junyan.xlsx
Untracked: data/Zenz Project overview cell counts etc.xlsx
Untracked: data/commonFiles/
Untracked: output/baseBH3.RData
Untracked: output/dataAnnexin.RData
Untracked: output/dynamicBH3.RData
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/_site.yml
Deleted: analysis/about.Rmd
Modified: analysis/index.Rmd
Deleted: analysis/license.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Preprocessing
Load data
Use baseline level from DBP profiling
Prepare sample background annotations
Association between PCs and clinical outcomes
PCA
PCA
Plot p value and hazard ratio 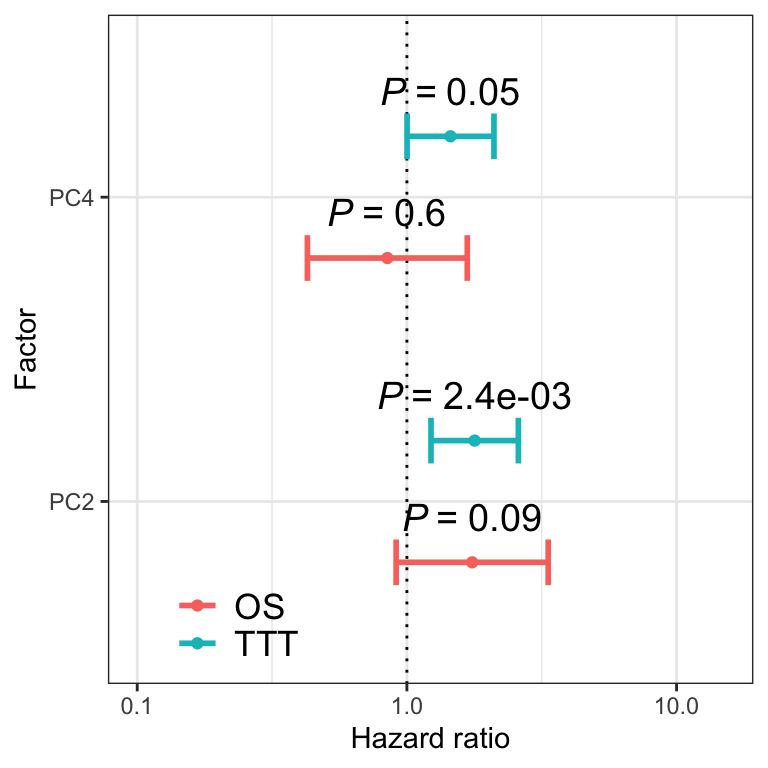
Kaplan-Meiler plots
KM plot for time to treatment (TTT) 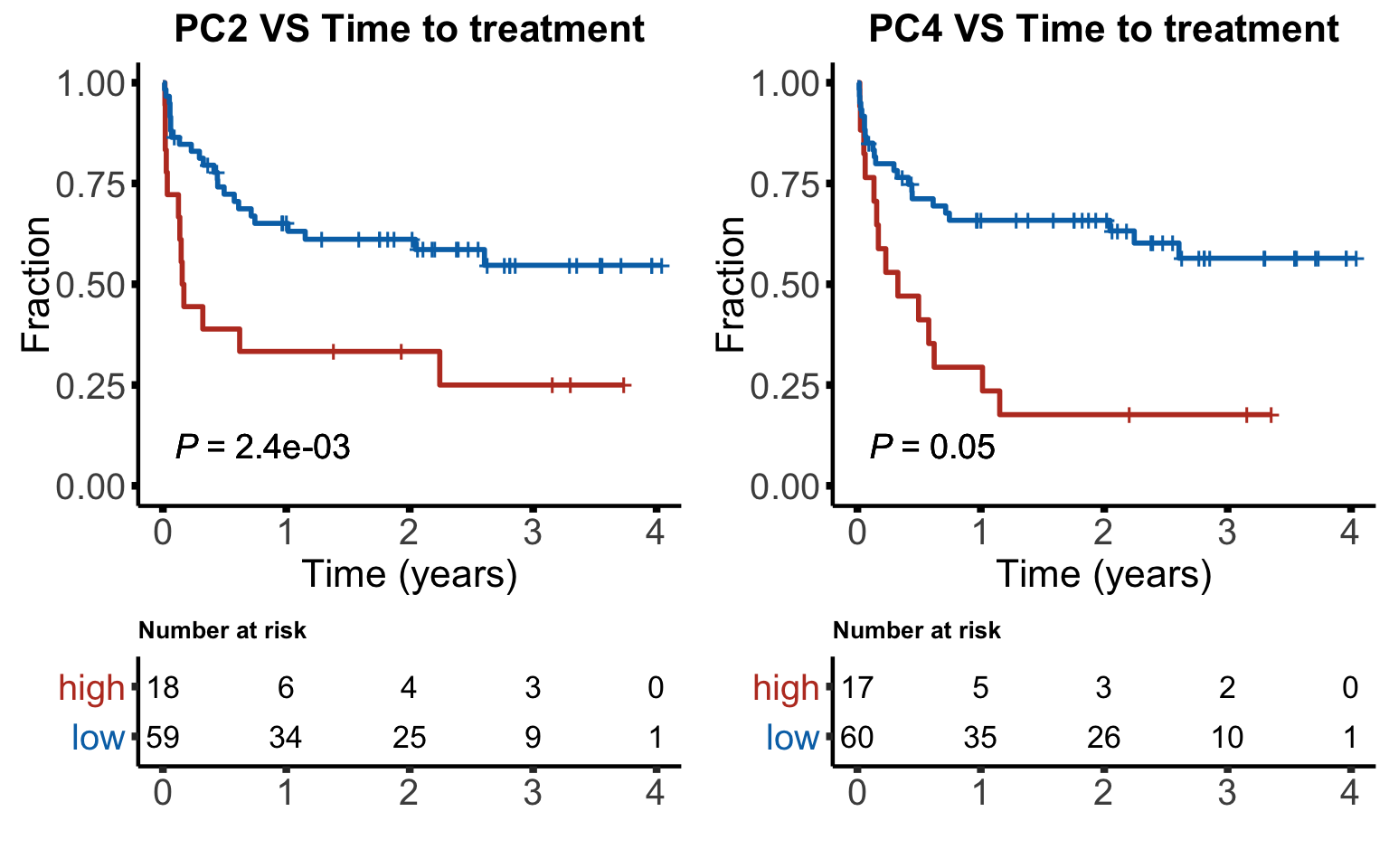
Correlation between PCs and CLL-PD
# A tibble: 6 x 2
# Groups: PC [6]
PC p.value
<chr> <dbl>
1 PC2 0.0000126
2 PC3 0.144
3 PC9 0.178
4 PC10 0.196
5 PC7 0.234
6 PC8 0.535 PC2 is associated with CLL-PD
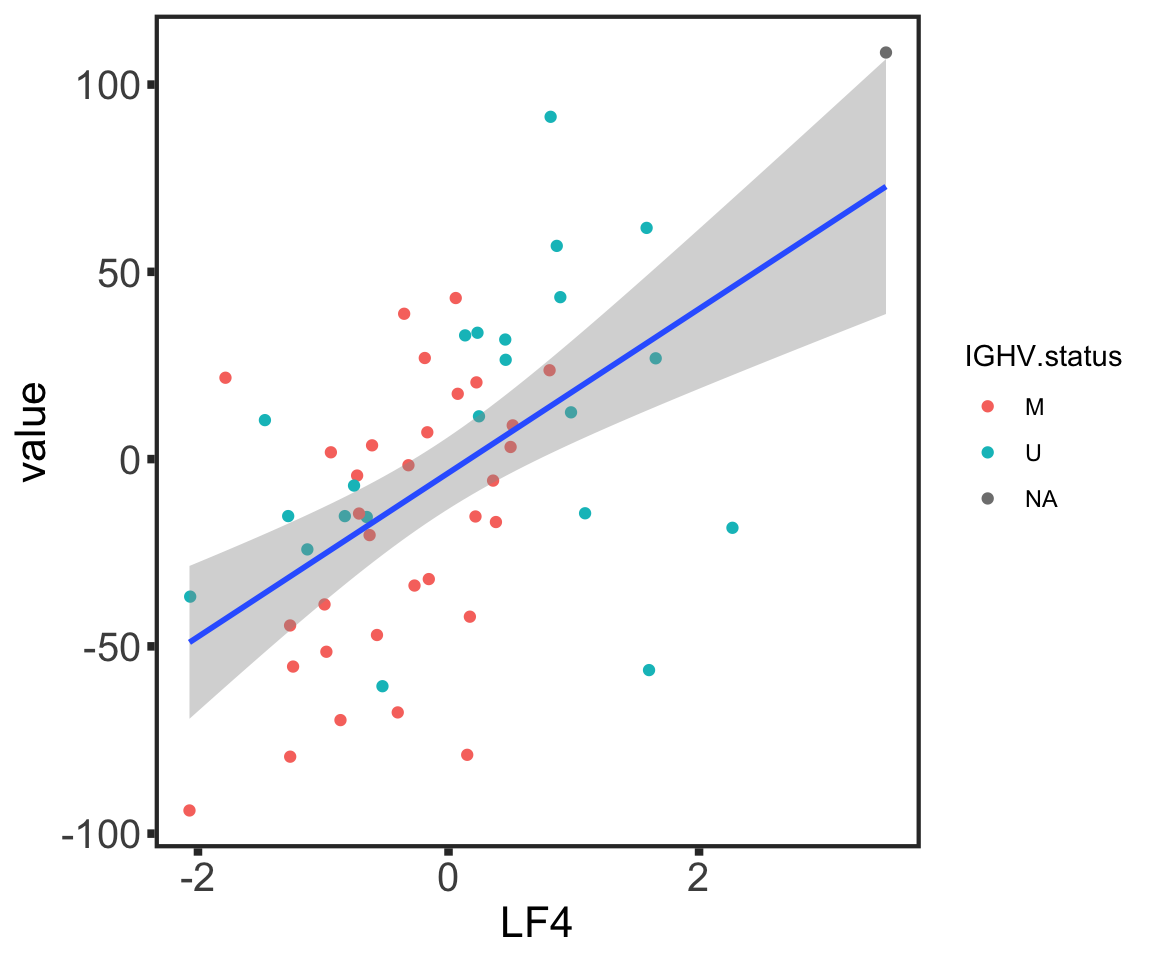
Anova Table (Type II tests)
Response: value
Sum Sq Df F value Pr(>F)
IGHV.status 3202 1 2.6452 0.110028
LF4 11487 1 9.4895 0.003326 **
Residuals 61734 51
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Select BH3 markers for outcome prediction
Prepare data
Univariate test
Table for TTT
Table for OS
Selection using multi-vairate model
Prepare data
Select based on C-index
TTT
Plot Harrel's C comparison 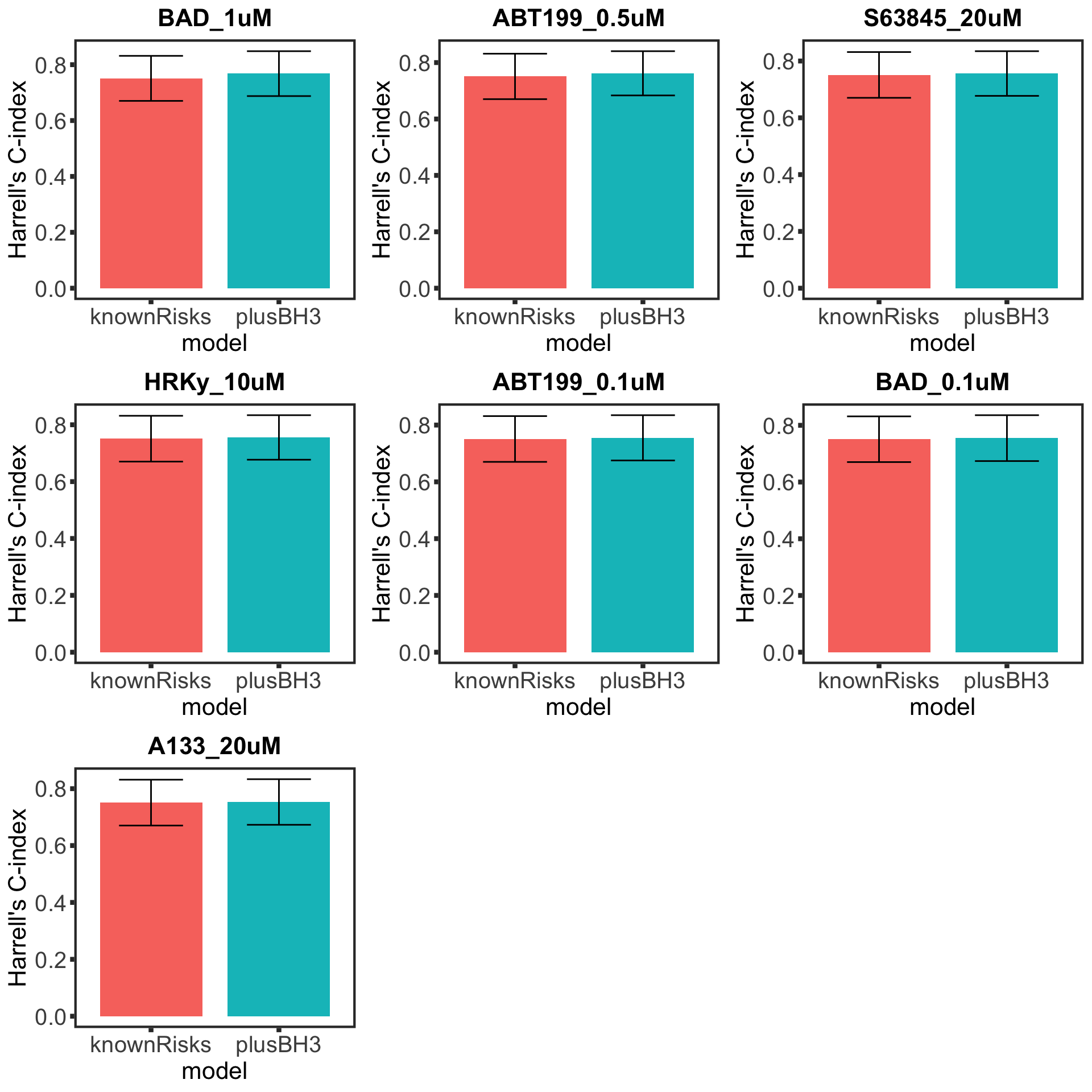
Forest plot of selected markers
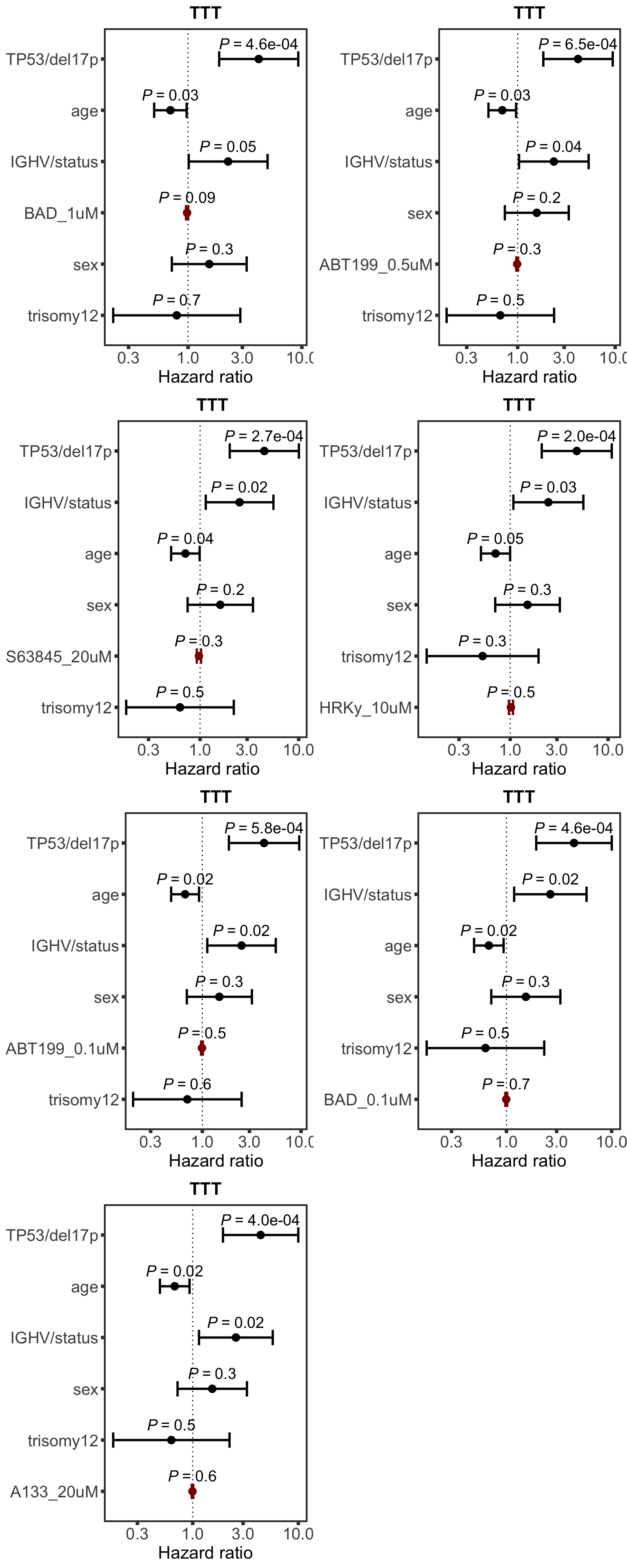 BH3 profile does not seem to provide additional information for predicting prognosis
BH3 profile does not seem to provide additional information for predicting prognosis
R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] latex2exp_0.4.0 forcats_0.5.0
[3] stringr_1.4.0 dplyr_1.0.0
[5] purrr_0.3.4 readr_1.3.1
[7] tidyr_1.1.0 tibble_3.0.3
[9] tidyverse_1.3.0 SummarizedExperiment_1.16.1
[11] DelayedArray_0.12.3 BiocParallel_1.20.1
[13] matrixStats_0.56.0 Biobase_2.46.0
[15] GenomicRanges_1.38.0 GenomeInfoDb_1.22.1
[17] IRanges_2.20.2 S4Vectors_0.24.4
[19] BiocGenerics_0.32.0 maxstat_0.7-25
[21] survminer_0.4.7 ggpubr_0.4.0
[23] ggplot2_3.3.2 survival_3.2-3
[25] jyluMisc_0.1.5 cowplot_1.0.0
[27] limma_3.42.2
loaded via a namespace (and not attached):
[1] readxl_1.3.1 backports_1.1.8 fastmatch_1.1-0
[4] drc_3.0-1 workflowr_1.6.2 igraph_1.2.5
[7] shinydashboard_0.7.1 splines_3.6.0 crosstalk_1.1.0.1
[10] TH.data_1.0-10 digest_0.6.25 htmltools_0.5.0
[13] fansi_0.4.1 gdata_2.18.0 magrittr_1.5
[16] cluster_2.1.0 openxlsx_4.1.5 modelr_0.1.8
[19] sandwich_2.5-1 piano_2.2.0 colorspace_1.4-1
[22] blob_1.2.1 rvest_0.3.5 haven_2.3.1
[25] xfun_0.15 crayon_1.3.4 RCurl_1.98-1.2
[28] jsonlite_1.7.0 zoo_1.8-8 glue_1.4.1
[31] gtable_0.3.0 zlibbioc_1.32.0 XVector_0.26.0
[34] car_3.0-8 abind_1.4-5 scales_1.1.1
[37] mvtnorm_1.1-1 DBI_1.1.0 relations_0.6-9
[40] rstatix_0.6.0 Rcpp_1.0.5 plotrix_3.7-8
[43] xtable_1.8-4 foreign_0.8-71 km.ci_0.5-2
[46] DT_0.14 htmlwidgets_1.5.1 httr_1.4.1
[49] fgsea_1.12.0 gplots_3.0.4 ellipsis_0.3.1
[52] mice_3.11.0 farver_2.0.3 pkgconfig_2.0.3
[55] dbplyr_1.4.4 utf8_1.1.4 labeling_0.3
[58] tidyselect_1.1.0 rlang_0.4.7 later_1.1.0.1
[61] munsell_0.5.0 cellranger_1.1.0 tools_3.6.0
[64] visNetwork_2.0.9 cli_2.0.2 generics_0.0.2
[67] broom_0.7.0 evaluate_0.14 fastmap_1.0.1
[70] yaml_2.2.1 knitr_1.29 fs_1.4.2
[73] zip_2.0.4 survMisc_0.5.5 caTools_1.18.0
[76] nlme_3.1-148 mime_0.9 slam_0.1-47
[79] xml2_1.3.2 compiler_3.6.0 rstudioapi_0.11
[82] curl_4.3 ggsignif_0.6.0 marray_1.64.0
[85] reprex_0.3.0 stringi_1.4.6 lattice_0.20-41
[88] Matrix_1.2-18 shinyjs_1.1 KMsurv_0.1-5
[91] vctrs_0.3.1 pillar_1.4.6 lifecycle_0.2.0
[94] data.table_1.12.8 bitops_1.0-6 httpuv_1.5.4
[97] R6_2.4.1 promises_1.1.1 KernSmooth_2.23-17
[100] gridExtra_2.3 rio_0.5.16 codetools_0.2-16
[103] MASS_7.3-51.6 gtools_3.8.2 exactRankTests_0.8-31
[106] assertthat_0.2.1 rprojroot_1.3-2 withr_2.2.0
[109] multcomp_1.4-13 GenomeInfoDbData_1.2.2 mgcv_1.8-31
[112] hms_0.5.3 grid_3.6.0 rmarkdown_2.3
[115] carData_3.0-4 git2r_0.27.1 sets_1.0-18
[118] shiny_1.5.0 lubridate_1.7.9