Some additional analysis for paper revision
Junyan Lu
2020 Sept. 3
Last updated: 2024-06-12
Checks: 6 1
Knit directory: BH3profiling/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200826) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 90ada8f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/manuscript_drugResponses_cache/
Ignored: analysis/manuscript_overview_cache/
Ignored: data/.DS_Store
Ignored: data/.Rapp.history
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/Annexin_rawInput.Rmd
Untracked: analysis/BH3_PCA.csv
Untracked: analysis/BH3baseline_analysis_fromBase.Rmd
Untracked: analysis/BH3baseline_analysis_fromDBP.Rmd
Untracked: analysis/BH3baseline_rawInput.Rmd
Untracked: analysis/BH3dynamic_rawInput.Rmd
Untracked: analysis/FigS5_RNA_num_associations.pdf
Untracked: analysis/analysisVenetoclax.Rmd
Untracked: analysis/based_pep_var.pdf
Untracked: analysis/clinialTrial_analysis.Rmd
Untracked: analysis/compareBaseline.Rmd
Untracked: analysis/compareBaseline.pdf
Untracked: analysis/deprecated/
Untracked: analysis/drugResponse_baseBH3_analysis.Rmd
Untracked: analysis/drugResponse_baseBH3_analysis_AUC copy.Rmd
Untracked: analysis/drugResponse_baseBH3_analysis_AUC.Rmd
Untracked: analysis/dynamicBH3_analysis.Rmd
Untracked: analysis/dynamicBH3_analysis_AUC.Rmd
Untracked: analysis/landscape_baseBH3_analysis.Rmd
Untracked: analysis/landscape_baseBH3_analysis_AUC.Rmd
Untracked: analysis/landscape_baseBH3_fromBase_analysis_AUC.Rmd
Untracked: analysis/manuscript_clinicalAnalysis.Rmd
Untracked: analysis/manuscript_drugResponses.Rmd
Untracked: analysis/manuscript_dynamicBH3.Rmd
Untracked: analysis/manuscript_dynamicBH3_old.Rmd
Untracked: analysis/manuscript_energyMetabolism.Rmd
Untracked: analysis/manuscript_overview.Rmd
Untracked: analysis/manuscript_overview_fromBaseBH3.Rmd
Untracked: analysis/manuscript_overview_singleConc.Rmd
Untracked: analysis/outcome_baseBH3_analysis.Rmd
Untracked: analysis/outcome_baseBH3_analysis_AUC.Rmd
Untracked: analysis/patBack.csv
Untracked: analysis/platePlot.pdf
Untracked: analysis/platePlot_dynamic.pdf
Untracked: analysis/predictOutcomes.Rmd
Untracked: analysis/pretreatment_effect_estiamtion.csv
Untracked: analysis/revisionAnalysis_ProtRNA_Genomic_correlations.Rmd
Untracked: analysis/tableS1_sampleSummary.csv
Untracked: analysis/treatTab.csv
Untracked: analysis/venePair_auc.pdf
Untracked: analysis/venePair_auc_norm.pdf
Untracked: analysis/venePair_auc_raw.pdf
Untracked: analysis/venePair_con.pdf
Untracked: analysis/venePair_conc_unorm.pdf
Untracked: code/utils.R
Untracked: data/13092020 Patient Outcome Analysis - IBR plus FCR - for Junyan.xlsx
Untracked: data/Compiled Baseline BH3 profile for Primary CLL_for Junyan.xlsx
Untracked: data/Data for Thorsten .xlsx
Untracked: data/Raw data Baseline for Junyan.xlsx
Untracked: data/Raw data DBP for Junyan.xlsx
Untracked: data/T. Zenz data for Junyan.xlsx
Untracked: data/Zenz Project overview cell counts etc.xlsx
Untracked: data/commonFiles/
Untracked: data/venetoclax_coculture.rds
Untracked: data/~$Compiled Baseline BH3 profile for Primary CLL_for Junyan.xlsx
Untracked: figures/
Untracked: manuscript/
Untracked: output/allData.RData
Untracked: output/baseBH3.RData
Untracked: output/dataAnnexin.RData
Untracked: output/dynamicBH3.RData
Untracked: prepareData.R
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/_site.yml
Deleted: analysis/about.Rmd
Modified: analysis/index.Rmd
Deleted: analysis/license.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Load and pre-process datasets
Define interested genes
Genes will be considered in the analysis
BCL-2 MCL-1 BCL-xL BFL-1 BAX BAK BIM PUMA
"BCL2" "MCL1" "BCL2L1" "BCL2A1" "BAX" "BAK1" "BCL2L11" "BBC3"
NOXA COX5A COX5B SOD1 SOD2
"PMAIP1" "COX5A" "COX5B" "SOD1" "SOD2" The first row shows the commonly used protein names and the second row shows the corresponding official gene symbols
BH3 profiling data
RNAseq data
BCL2 MCL1 BCL2L1 BCL2A1 BAX BAK1 BCL2L11 BBC3 PMAIP1 COX5A
TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
COX5B SOD1 SOD2
TRUE TRUE TRUE [1] 13 71Proteomics
BCL2 MCL1 BCL2L1 BCL2A1 BAX BAK1 BCL2L11 BBC3 PMAIP1 COX5A
TRUE FALSE FALSE FALSE TRUE FALSE TRUE FALSE FALSE TRUE
COX5B SOD1 SOD2
TRUE FALSE TRUE [1] 6 91Genomics
Prepare patient genomic background
[1] "IGHV.status" "del11q" "del13q" "del17p" "trisomy12"
[6] "NOTCH1" "SF3B1" "TP53" Correlation between the baseline mRNA expression of the interested genes and mutations in CLL
Result table
Plot associations with 10% FDR
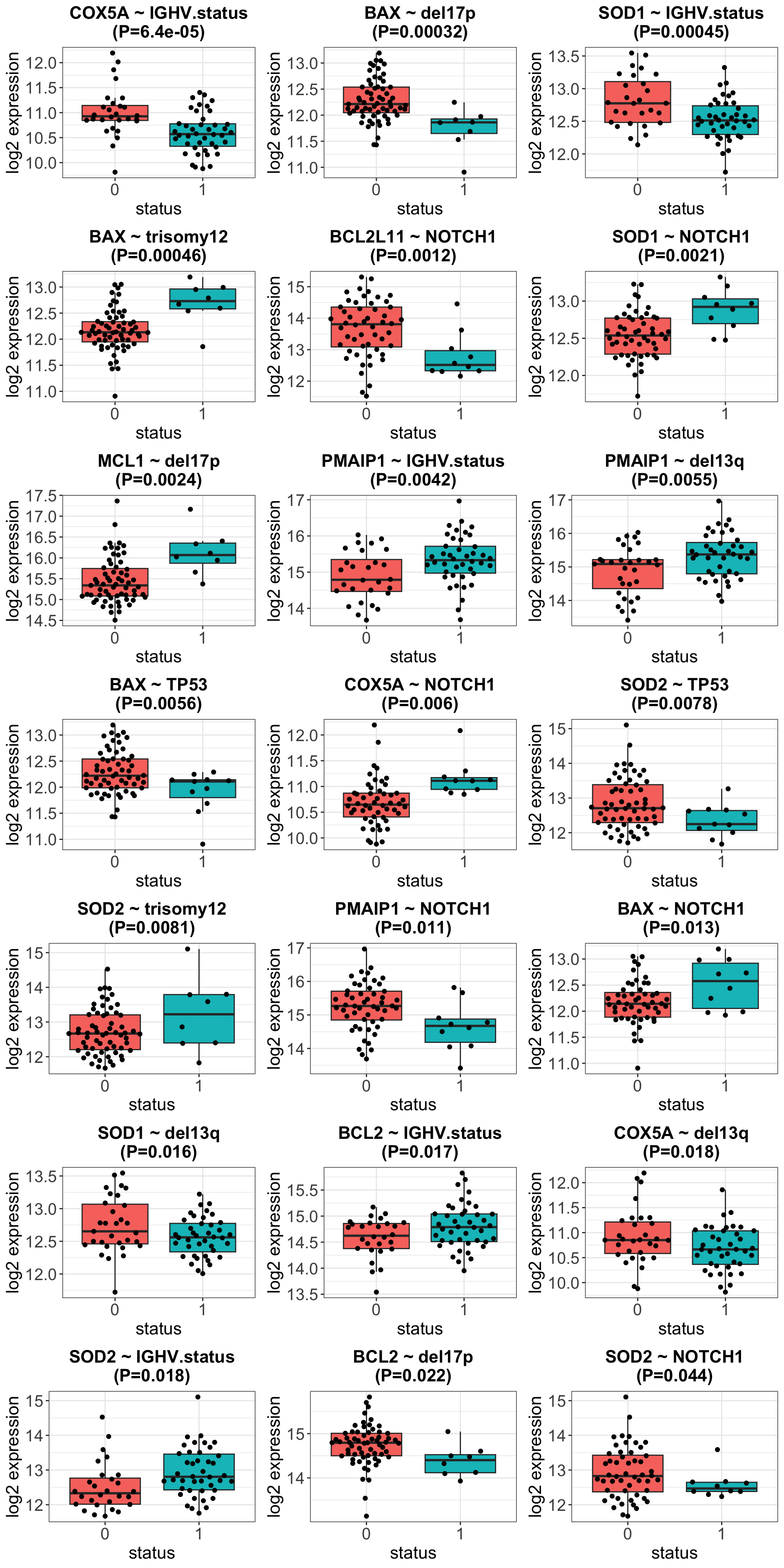
Correlation between the protein expressions of the interested genes and mutations in CLL
Result table
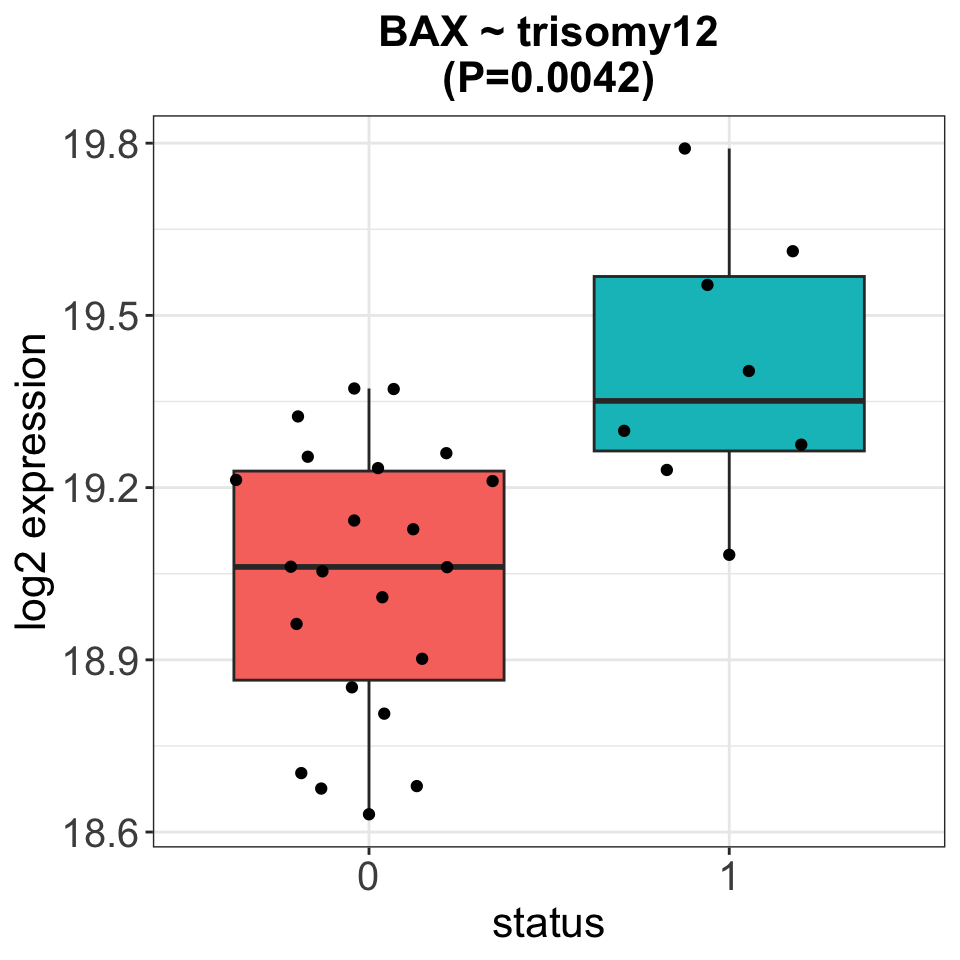
Only the associations between BAX and trisomy12 passed 10% FDR
Plot associations with 10% FDR
Differential expression of the interested genes before and after ex-vivo Ibrutinib treatment
Load dataset
Preprocessing
Subset for DMSO and Ibr
Subset for interested genes
[1] 13 187Differential expression between Ibr treatment and DMSO
Plot associations with 10% FDR
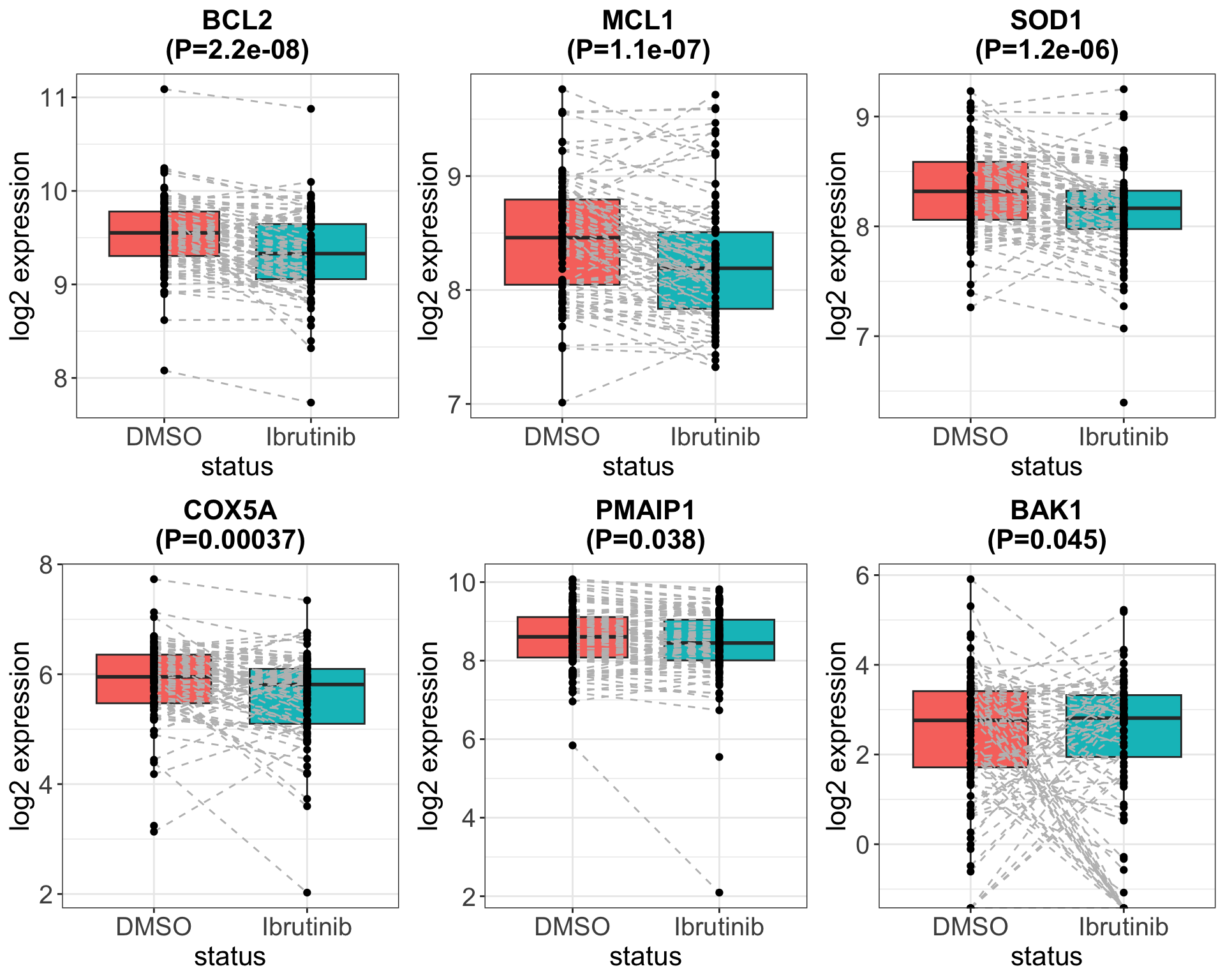
Wether the expression of interested genes before and after ex-vivo ibrutinib treatment associated with ibrutinib responses?
Prepare drug response profile
Prepare gene expression
Association test
Table of significant associations (10%)
In the treatment column:
- Ibrutinib_48h means the gene expression after
Ibrutinib treatment for 48 hours
- DMSO_48h means the gene expression after 48h
incubation with just DMSO
- FoldChange means the log fold change between
Ibrutinib and DMSO after 48 hourse
Note that in this experiment, we don’t have the real baseline (0 hour)
Plot the significant associations
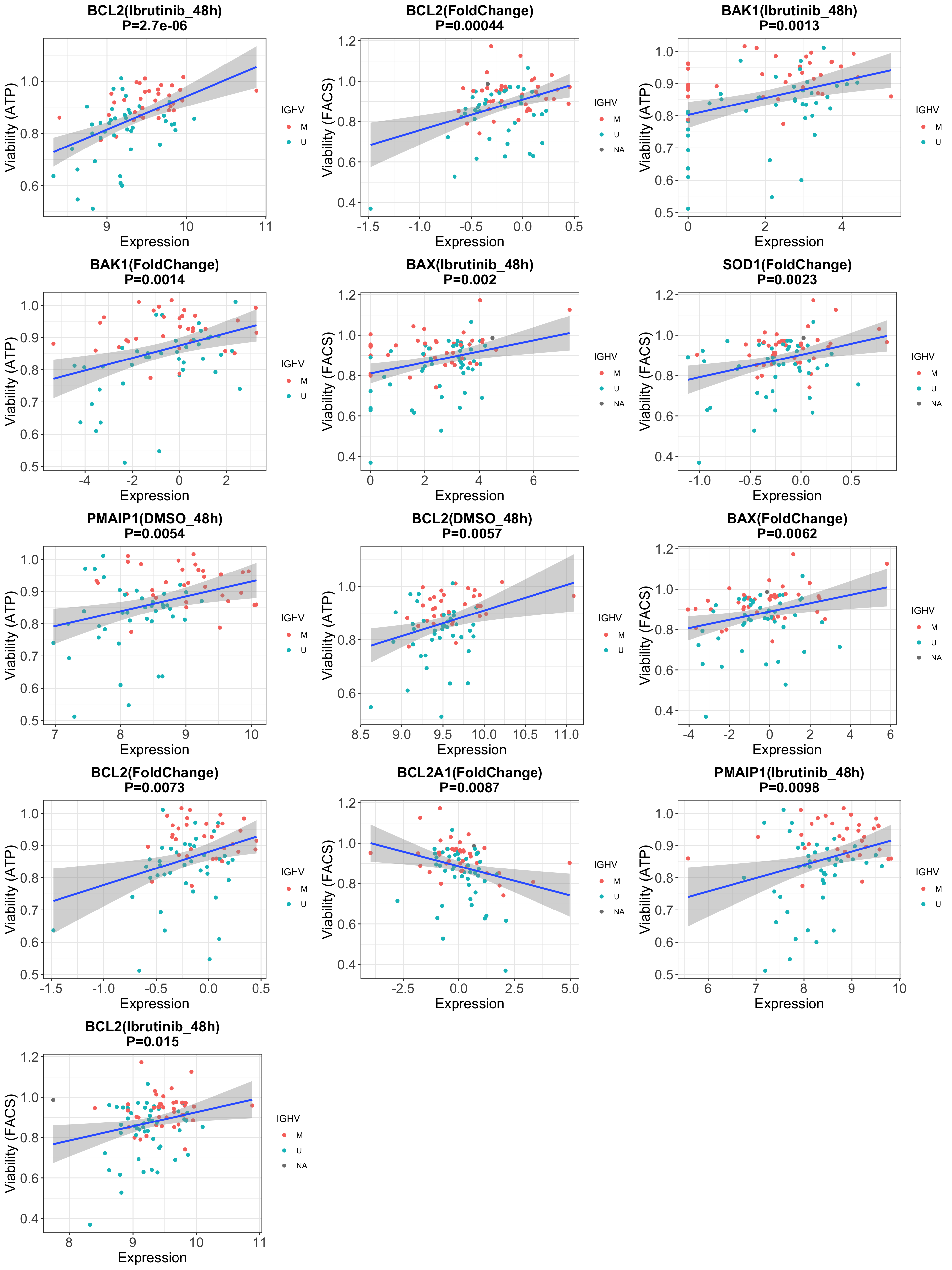
Wether the expression of interested genes before and after ex-vivo ibrutinib treatment associated with BH3 profiling data?
Prepare drug response profile
Association test
Table of significant associations (10%)
Plot the significant associations
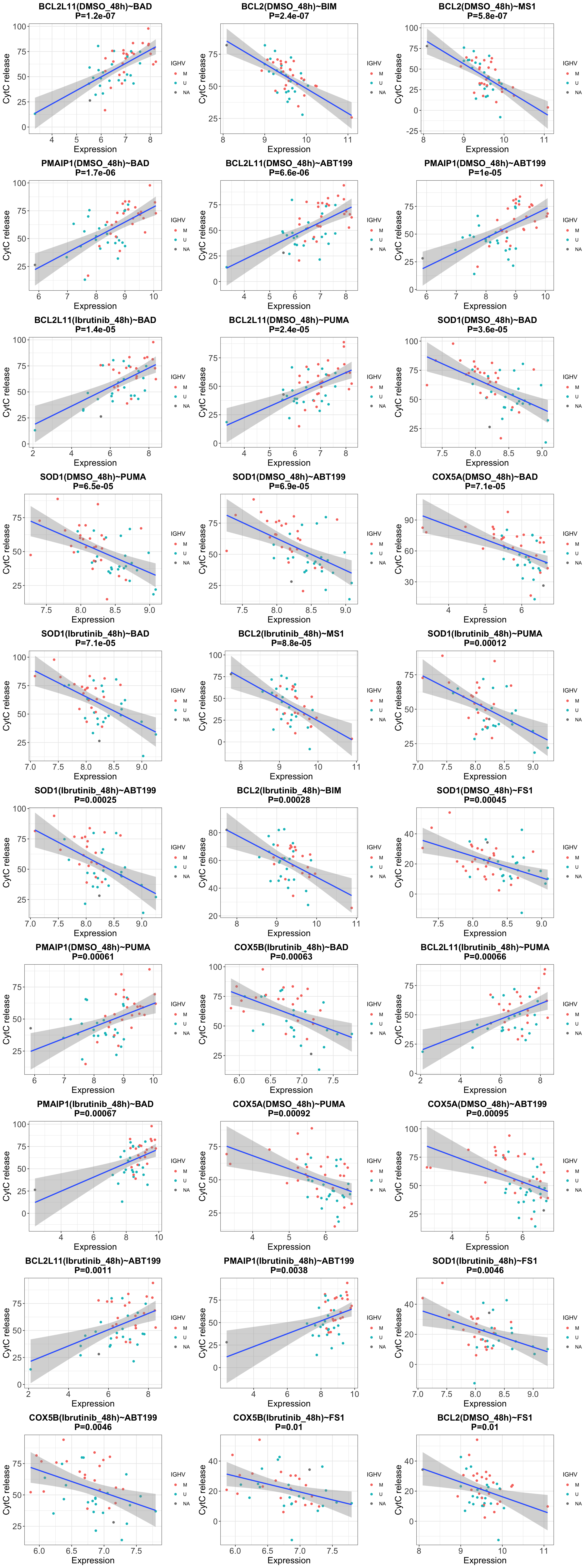 It’s interesting to see that the gene expression after 48h DMSO
incubation show stronger association to BH3 profile than the real
baseline gene expression (0h) we used in the manuscript
It’s interesting to see that the gene expression after 48h DMSO
incubation show stronger association to BH3 profile than the real
baseline gene expression (0h) we used in the manuscript
This could be because the BH3 profiling data we are using is
also the BH3 profile after 48h DMSO incubation
You can see from the below longitudinal gene expression profile,
some of those genes show strong changes during culturing in
DMSO
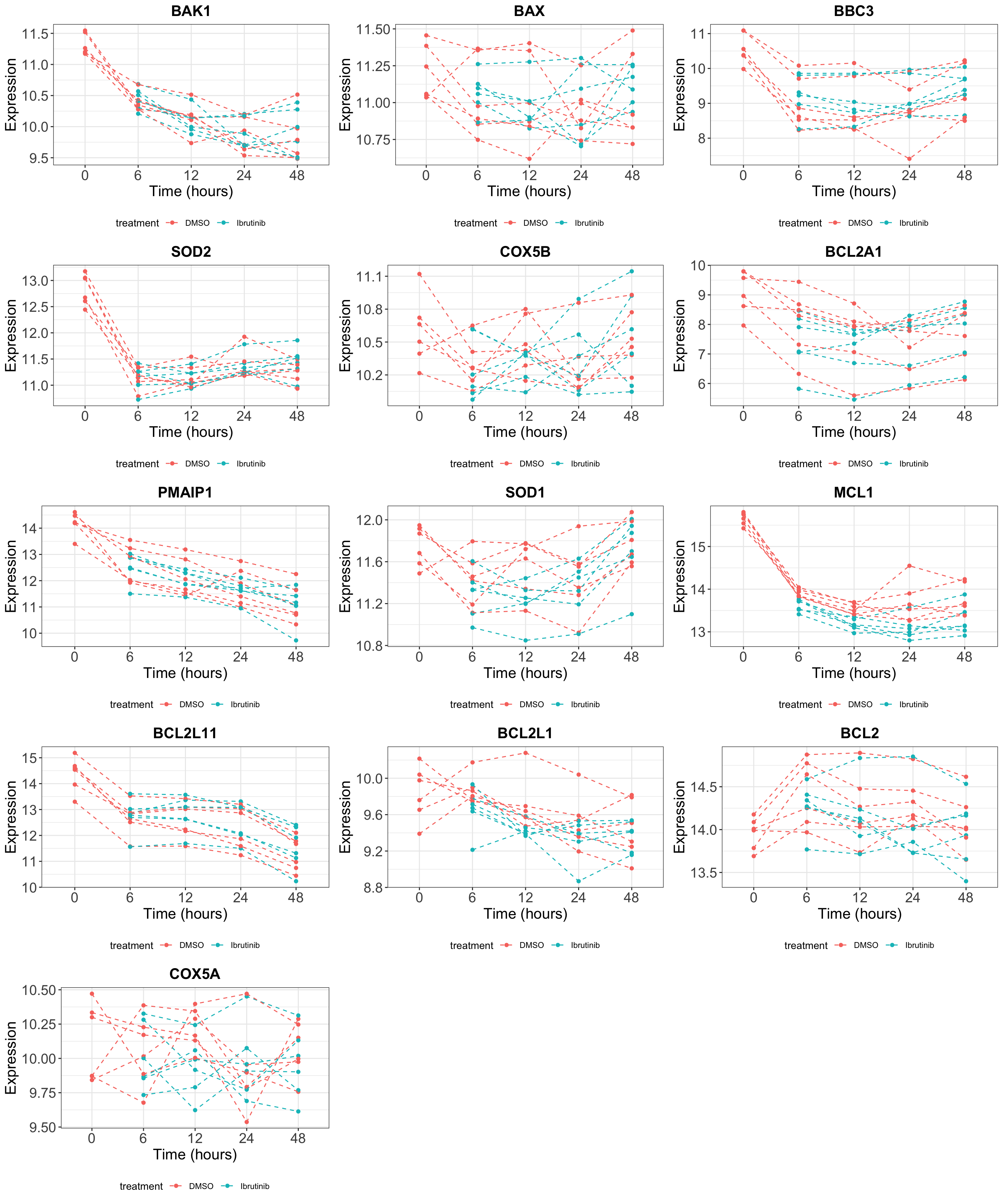
Look at the longitudinal gene expression data
Visualize the time-course expression of the interested genes in DMSO and Ibrutinib treated samples
Correlate the protein expression with in-vivo Ibr treated patients (a smaller cohort from Heidelberg)
Load dataset
Subset for interested genes
[1] 6 10Result table
Plot associations with 10% FDR
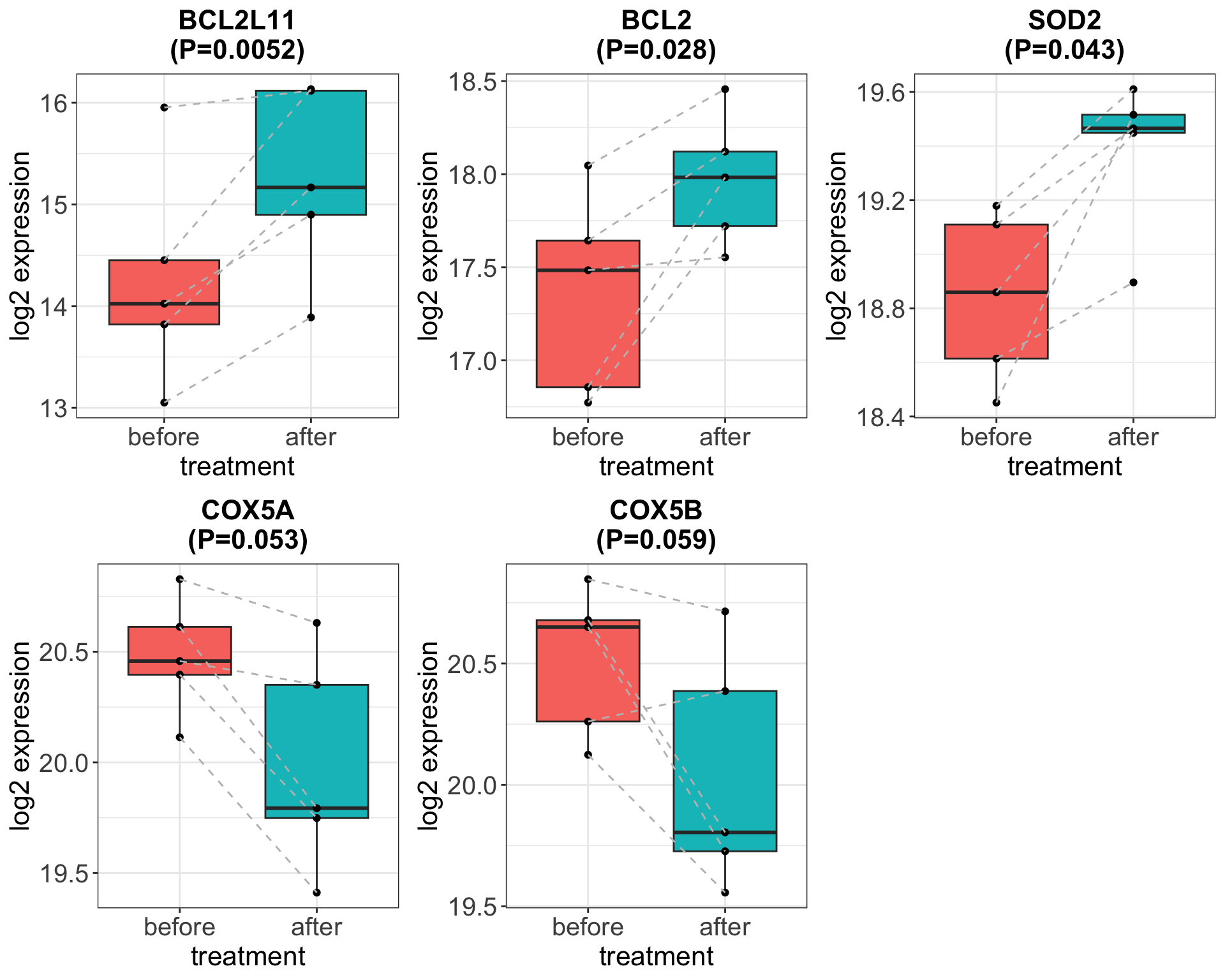
Contrary to the gene expression after ex-vivo Ibr treatment, BCL2 protein level increased after in-vivo ibrutinib treatment
Correlate the protein expression with in-vivo Ibr treated patients (look at a larger cohort from Zurich)
Load dataset
Selected appropriate samples
[1] 9 30Differential expression
Result table
Plot associations with 10% FDR
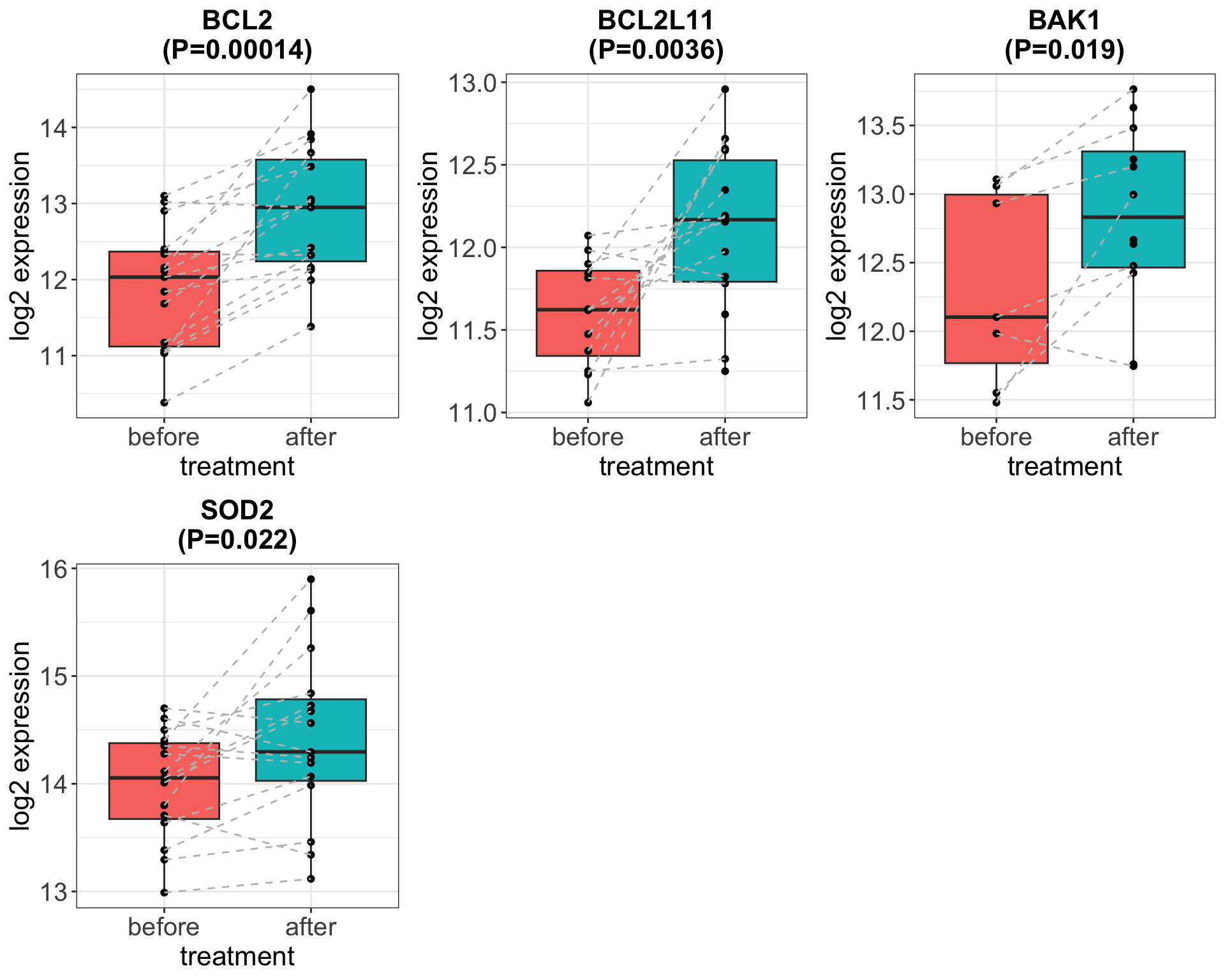 The trend is similar to the smaller cohort from
Heidelberg
The trend is similar to the smaller cohort from
Heidelberg
Correlate the gene expression with in-vivo Ibr treated patients (those are the sample samples as the ones above)
Load dataset
Selected appropriate samples
[1] 12 70Differential expression
Plot associations with 10% FDR
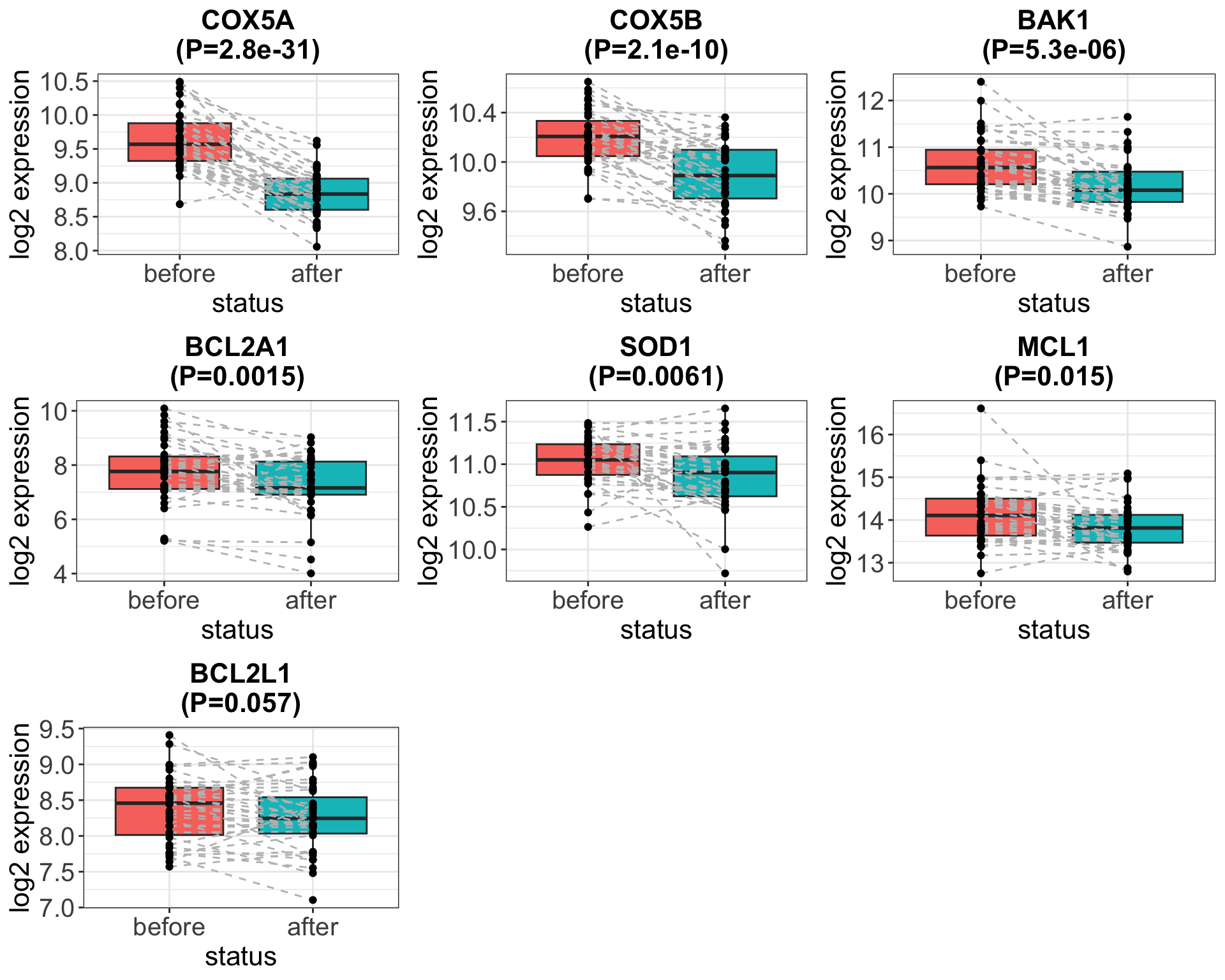 It seems the trend on mRNA level is not consistent with the
trend on protein level..
It seems the trend on mRNA level is not consistent with the
trend on protein level..
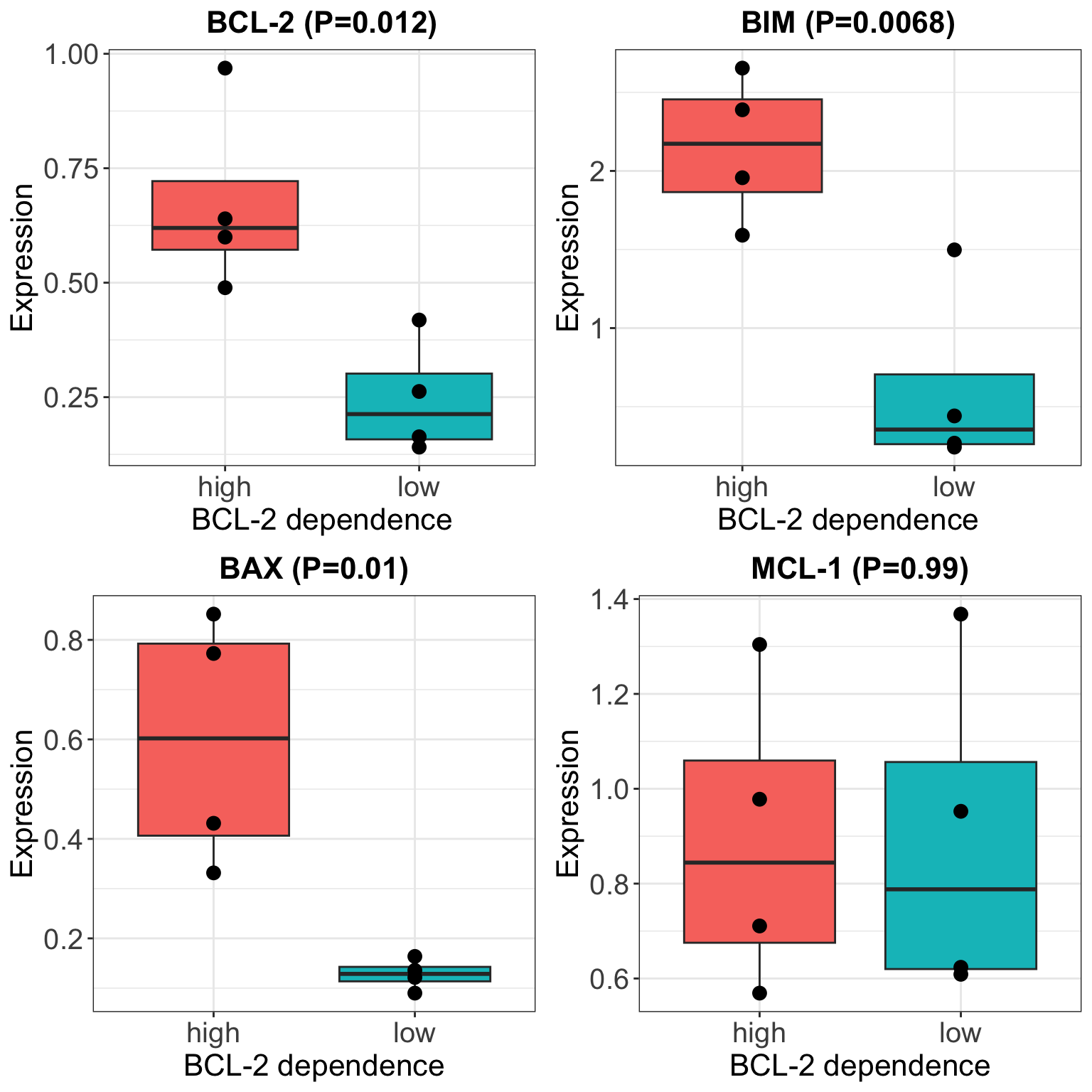
Analysis of Stephen’s Western Blot data
T-test
Boxplot
R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] latex2exp_0.9.4 forcats_0.5.1
[3] stringr_1.4.1 dplyr_1.1.4.9000
[5] purrr_0.3.4 readr_2.1.2
[7] tidyr_1.2.0 tibble_3.2.1
[9] ggplot2_3.4.1 tidyverse_1.3.2
[11] DESeq2_1.36.0 SummarizedExperiment_1.26.1
[13] Biobase_2.56.0 MatrixGenerics_1.8.1
[15] matrixStats_0.62.0 GenomicRanges_1.48.0
[17] GenomeInfoDb_1.32.2 IRanges_2.30.0
[19] S4Vectors_0.34.0 BiocGenerics_0.42.0
[21] jyluMisc_0.1.5
loaded via a namespace (and not attached):
[1] utf8_1.2.4 shinydashboard_0.7.2 tidyselect_1.2.1
[4] RSQLite_2.2.15 AnnotationDbi_1.58.0 htmlwidgets_1.5.4
[7] grid_4.2.0 BiocParallel_1.30.3 maxstat_0.7-25
[10] munsell_0.5.0 codetools_0.2-18 DT_0.23
[13] withr_3.0.0 colorspace_2.0-3 highr_0.9
[16] knitr_1.39 proDA_1.10.0 rstudioapi_0.13
[19] ggsignif_0.6.3 labeling_0.4.2 git2r_0.30.1
[22] slam_0.1-50 GenomeInfoDbData_1.2.8 KMsurv_0.1-5
[25] farver_2.1.1 bit64_4.0.5 rprojroot_2.0.3
[28] vctrs_0.6.5 generics_0.1.3 TH.data_1.1-1
[31] xfun_0.31 sets_1.0-21 R6_2.5.1
[34] ggbeeswarm_0.6.0 locfit_1.5-9.6 bitops_1.0-7
[37] cachem_1.0.6 fgsea_1.22.0 DelayedArray_0.22.0
[40] assertthat_0.2.1 promises_1.2.0.1 scales_1.2.0
[43] multcomp_1.4-19 googlesheets4_1.0.0 beeswarm_0.4.0
[46] gtable_0.3.0 extraDistr_1.9.1 sandwich_3.0-2
[49] workflowr_1.7.0 rlang_1.1.3 genefilter_1.78.0
[52] splines_4.2.0 rstatix_0.7.0 gargle_1.2.0
[55] broom_1.0.0 yaml_2.3.5 abind_1.4-5
[58] modelr_0.1.8 crosstalk_1.2.0 backports_1.4.1
[61] httpuv_1.6.6 tools_4.2.0 relations_0.6-12
[64] ellipsis_0.3.2 gplots_3.1.3 jquerylib_0.1.4
[67] RColorBrewer_1.1-3 Rcpp_1.0.9 visNetwork_2.1.0
[70] zlibbioc_1.42.0 RCurl_1.98-1.7 ggpubr_0.4.0
[73] cowplot_1.1.1 zoo_1.8-10 haven_2.5.0
[76] cluster_2.1.3 exactRankTests_0.8-35 fs_1.5.2
[79] magrittr_2.0.3 data.table_1.14.8 reprex_2.0.1
[82] survminer_0.4.9 googledrive_2.0.0 mvtnorm_1.1-3
[85] hms_1.1.1 shinyjs_2.1.0 mime_0.12
[88] evaluate_0.15 xtable_1.8-4 XML_3.99-0.10
[91] readxl_1.4.0 gridExtra_2.3 compiler_4.2.0
[94] KernSmooth_2.23-20 crayon_1.5.2 htmltools_0.5.4
[97] mgcv_1.8-40 later_1.3.0 tzdb_0.3.0
[100] geneplotter_1.74.0 lubridate_1.8.0 DBI_1.1.3
[103] dbplyr_2.2.1 MASS_7.3-58 Matrix_1.5-4
[106] car_3.1-0 cli_3.6.2 marray_1.74.0
[109] parallel_4.2.0 igraph_1.3.4 pkgconfig_2.0.3
[112] km.ci_0.5-6 piano_2.12.0 xml2_1.3.3
[115] annotate_1.74.0 vipor_0.4.5 bslib_0.4.1
[118] XVector_0.36.0 drc_3.0-1 rvest_1.0.2
[121] digest_0.6.30 Biostrings_2.64.0 rmarkdown_2.14
[124] cellranger_1.1.0 fastmatch_1.1-3 survMisc_0.5.6
[127] shiny_1.7.4 gtools_3.9.3 nlme_3.1-158
[130] lifecycle_1.0.4 jsonlite_1.8.3 carData_3.0-5
[133] limma_3.52.2 fansi_1.0.6 pillar_1.9.0
[136] lattice_0.20-45 KEGGREST_1.36.3 fastmap_1.1.0
[139] httr_1.4.3 plotrix_3.8-2 survival_3.4-0
[142] glue_1.7.0 png_0.1-7 bit_4.0.4
[145] stringi_1.7.8 sass_0.4.2 blob_1.2.3
[148] caTools_1.18.2 memoise_2.0.1