Exploratory data analysis and QA
Last updated: 2022-11-17
Checks: 5 1
Knit directory:
LungCancer_SotilloLab/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221103) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the
results is critical for reproducibility. To start using Git, open the
Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full
reproducibility benefits of using workflowr, please see
?wflow_start.
Load packages and dataset
#package
library(SummarizedExperiment)
library(MultiAssayExperiment)
library(proDA)
library(tidyverse)
source("../code/utils.R")
knitr::opts_chunk$set(warning = FALSE, message = FALSE, autodep = TRUE)Load processed data
load("../output/processedData.RData")Phosphoproteomic
Preprocessing
Use different normalization method
#variance stabilizing normalization
ppe.vst <- preprocessPhos(maeData, missCut = 0.5, transform = "vst")[1] "Number of proteins and samples:"
[1] 3787 96#log2 + median normalization
ppe.log2Med <- preprocessPhos(maeData, missCut = 0.5, transform = "log2", normalize = TRUE)[1] "Number of proteins and samples:"
[1] 3787 96#only log2 transformation
ppe.log2Only <- preprocessPhos(maeData, missCut = 0.5, transform ="log2", normalize = FALSE)[1] "Number of proteins and samples:"
[1] 3787 96#log2 transformation + normalization based on precursur quantity
ppe.pre <- preprocessPhos(maeData, missCut = 0.5, transform ="log2", normalize = TRUE, usePrecursor = TRUE)[1] "Number of proteins and samples:"
[1] 3787 96Mean SD plots
VST
vsn::meanSdPlot(assay(ppe.vst))
Log2+median
vsn::meanSdPlot(assay(ppe.log2Med))
log2 only
vsn::meanSdPlot(assay(ppe.log2Only))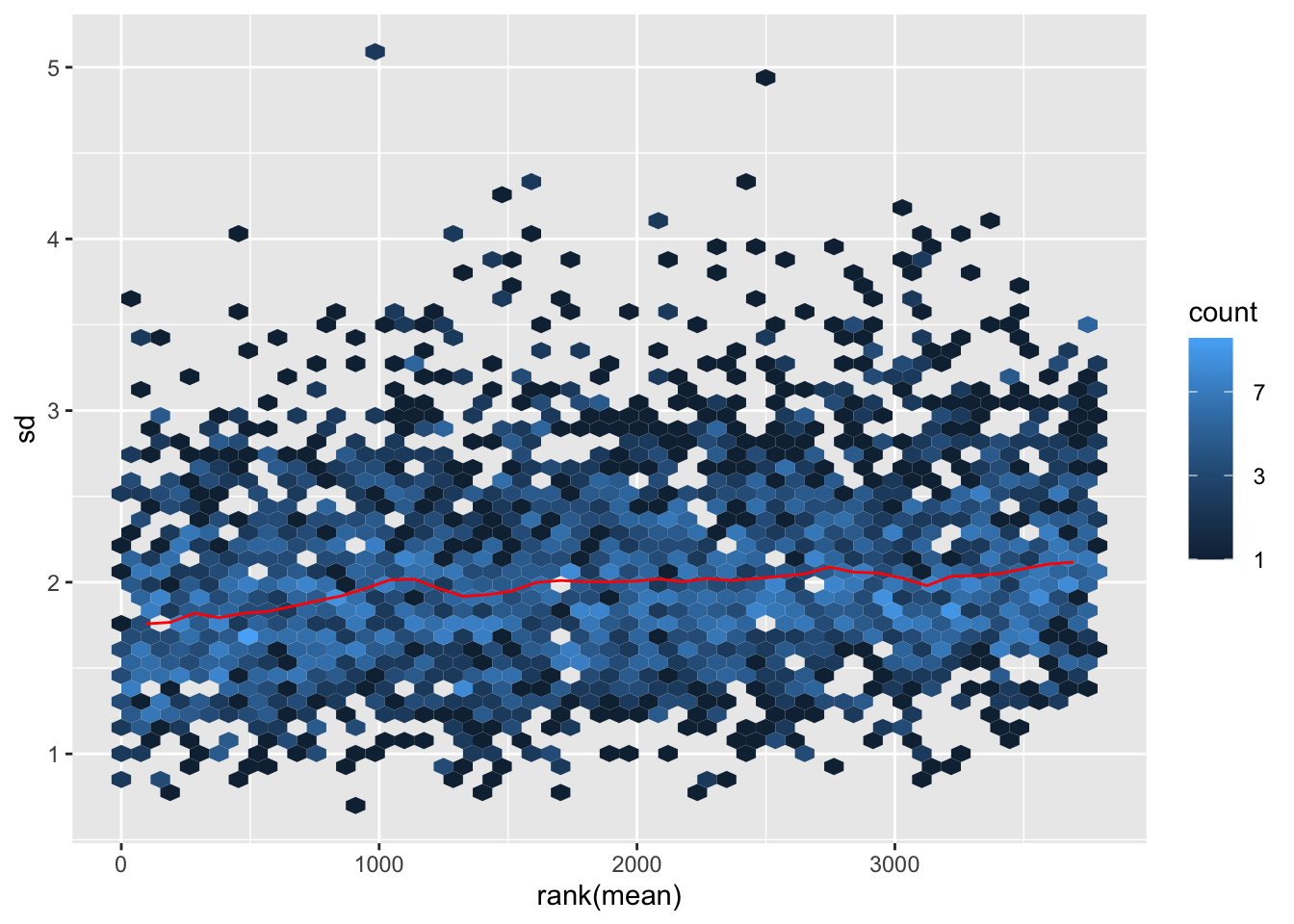 #### Use precursor
#### Use precursor
vsn::meanSdPlot(assay(ppe.pre))
Distribution after normalization
VST
countMat <- assay(ppe.vst)
annoTab <- colData(ppe.vst)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
log2+median
countMat <- assay(ppe.log2Med)
annoTab <- colData(ppe.log2Med)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))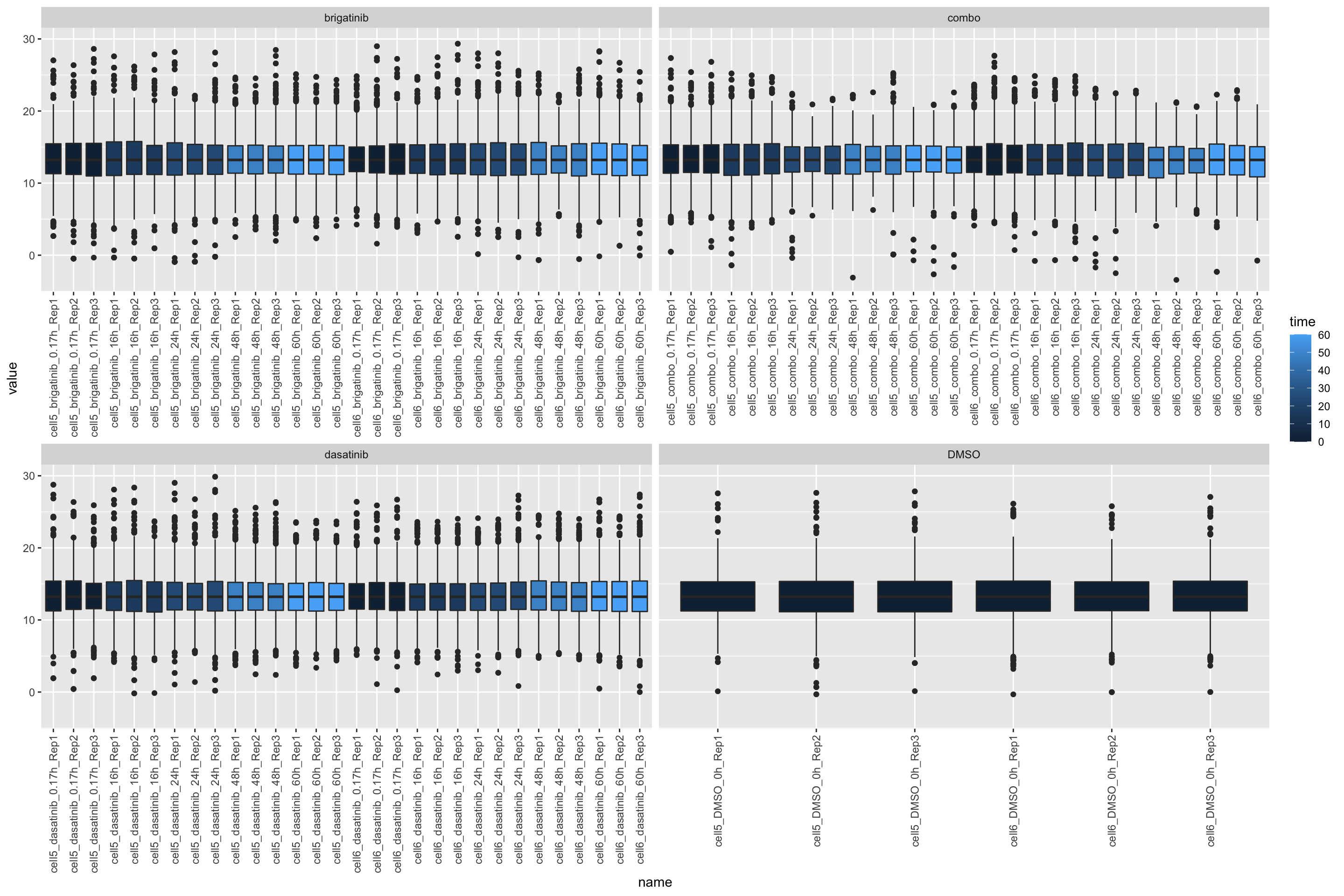 #### only transformation
#### only transformation
countMat <- assay(ppe.log2Only)
annoTab <- colData(ppe.log2Only)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
use precursor
countMat <- assay(ppe.pre)
annoTab <- colData(ppe.pre)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
PCA
VST
exprMat <- assays(ppe.vst)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.vst) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2+median
exprMat <- assays(ppe.log2Med)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Med) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2 only
exprMat <- assays(ppe.log2Only)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Only) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Use precursor
exprMat <- assays(ppe.pre)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.pre) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
VST
colAnno <- colData(ppe.vst)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2+median
colAnno <- colData(ppe.log2Med)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2 only
colAnno <- colData(ppe.log2Only)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
precursor
colAnno <- colData(ppe.pre)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Remove one potential outlier
maeSub <- maeData[, maeData$sample != "cell5_combo_24h_Rep2"]
#variance stabilizing normalization
ppe.vst <- preprocessPhos(maeSub, missCut = 0.5, transform = "vst")[1] "Number of proteins and samples:"
[1] 3879 95#log2 + median normalization
ppe.log2Med <- preprocessPhos(maeSub, missCut = 0.5, transform = "log2", normalize = TRUE)[1] "Number of proteins and samples:"
[1] 3879 95#only log2 transformation
ppe.log2Only <- preprocessPhos(maeSub, missCut = 0.5, transform ="log2", normalize = FALSE)[1] "Number of proteins and samples:"
[1] 3879 95#log2 transformation + normalization based on precursur quantity
ppe.pre <- preprocessPhos(maeData, missCut = 0.5, transform ="log2", normalize = TRUE, usePrecursor = TRUE)[1] "Number of proteins and samples:"
[1] 3787 96PCA
VST
exprMat <- assays(ppe.vst)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.vst) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2+median
exprMat <- assays(ppe.log2Med)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Med) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2 only
exprMat <- assays(ppe.log2Only)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Only) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Use precursor
exprMat <- assays(ppe.pre)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.pre) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
VST
colAnno <- colData(ppe.vst)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2+median
colAnno <- colData(ppe.log2Med)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2 only
colAnno <- colData(ppe.log2Only)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
precursor
colAnno <- colData(ppe.pre)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Compare the reproducibiliy of replicates when using different normalization methods
Function to calculate reproducibility of replicates
getSdTab <- function(x) {
sd <- jyluMisc::sumToTidy(x) %>%
group_by(sampleCondi, rowID) %>%
summarise(sdVal = sd(Intensity,na.rm=TRUE), meanVal = mean(Intensity,na.rm=TRUE)) %>%
filter(!is.na(sdVal)) %>% mutate(meanRnk = order(meanVal))
}sdTab.vst <- getSdTab(ppe.vst) %>% mutate(norm = "vst")
sdTab.log2Med <- getSdTab(ppe.log2Med) %>% mutate(norm = "log2Med")
sdTab.log2Only <- getSdTab(ppe.log2Only) %>% mutate(norm = "log2Only")
sdTab.pre <- getSdTab(ppe.pre) %>% mutate(norm = "precursor")
sumTab <- bind_rows(sdTab.vst, sdTab.log2Med, sdTab.log2Only, sdTab.pre) Add annotations
colTab <- colData(ppe.vst) %>% as_tibble() %>%
distinct(sampleCondi,.keep_all = TRUE)
sumTab <- left_join(sumTab, colTab)Overall distribution
ggplot(sumTab, aes(x=sdVal, fill = norm)) +
geom_histogram(position = "identity", alpha=0.5, color = "grey50") +
xlim(0,6)
Per sample
ggplot(sumTab, aes(x=sampleCondi, y = sdVal, fill = norm)) +
geom_violin() +
facet_wrap(~drug, scale="free_x")+
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
Mean versus SD
ggplot(sumTab, aes(x=meanVal, y=sdVal)) +
geom_hex() + geom_smooth() +
facet_wrap(~norm, ncol=1)
Rank of sd for each normalization method
ordTab <- group_by(sumTab, sampleCondi, rowID) %>%
mutate(index = order(sdVal))
ggplot(ordTab, aes(x=index, fill = norm)) +
geom_bar(position = "dodge", alpha=0.5, color = "grey50") 
For each sample, which normalization method gives the best reproduciblity for replicates
ordPerTab <- arrange(ordTab, index) %>% distinct(sampleCondi, drug,.keep_all = TRUE) %>%
group_by(sampleCondi, norm, drug) %>% summarise(n=length(rowID))
ggplot(ordPerTab, aes(x=sampleCondi, y = n, fill = norm)) +
geom_bar(stat="identity")+
facet_wrap(~drug, scale = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))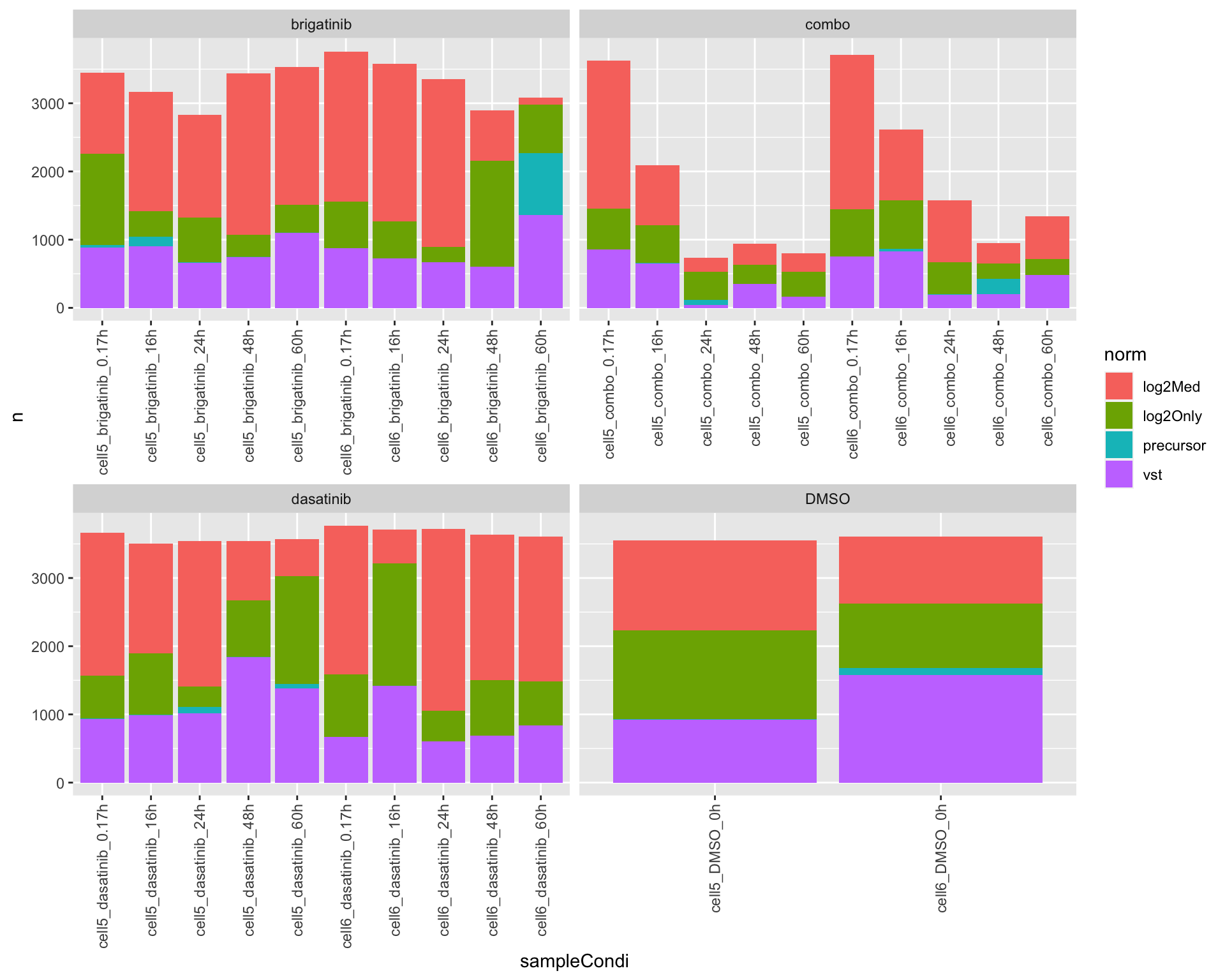 It seems log2 + median scaling can get the best reproducibility among
replicates. No normalization is the worst.
It seems log2 + median scaling can get the best reproducibility among
replicates. No normalization is the worst.
Proteomic
Preprocessing
Use different normalization method
#variance stabilizing normalization
ppe.vst <- preprocessProteome(maeData, missCut = 0.5, transform = "vst")[1] "Number of proteins and samples:"
[1] 7608 96#log2 + median normalization
ppe.log2Med <- preprocessProteome(maeData, missCut = 0.5, transform = "log2", normalize = TRUE)[1] "Number of proteins and samples:"
[1] 7608 96#only log2 transformation
ppe.log2Only <- preprocessProteome(maeData, missCut = 0.5, transform ="log2", normalize = FALSE)[1] "Number of proteins and samples:"
[1] 7608 96#log2 transformation + normalization based on precursur quantity
ppe.pre <- preprocessProteome(maeData, missCut = 0.5, transform ="log2", normalize = TRUE, usePrecursor = TRUE)[1] "Number of proteins and samples:"
[1] 7608 96Mean SD plots
VST
vsn::meanSdPlot(assay(ppe.vst))
Log2+median
vsn::meanSdPlot(assay(ppe.log2Med))
log2 only
vsn::meanSdPlot(assay(ppe.log2Only))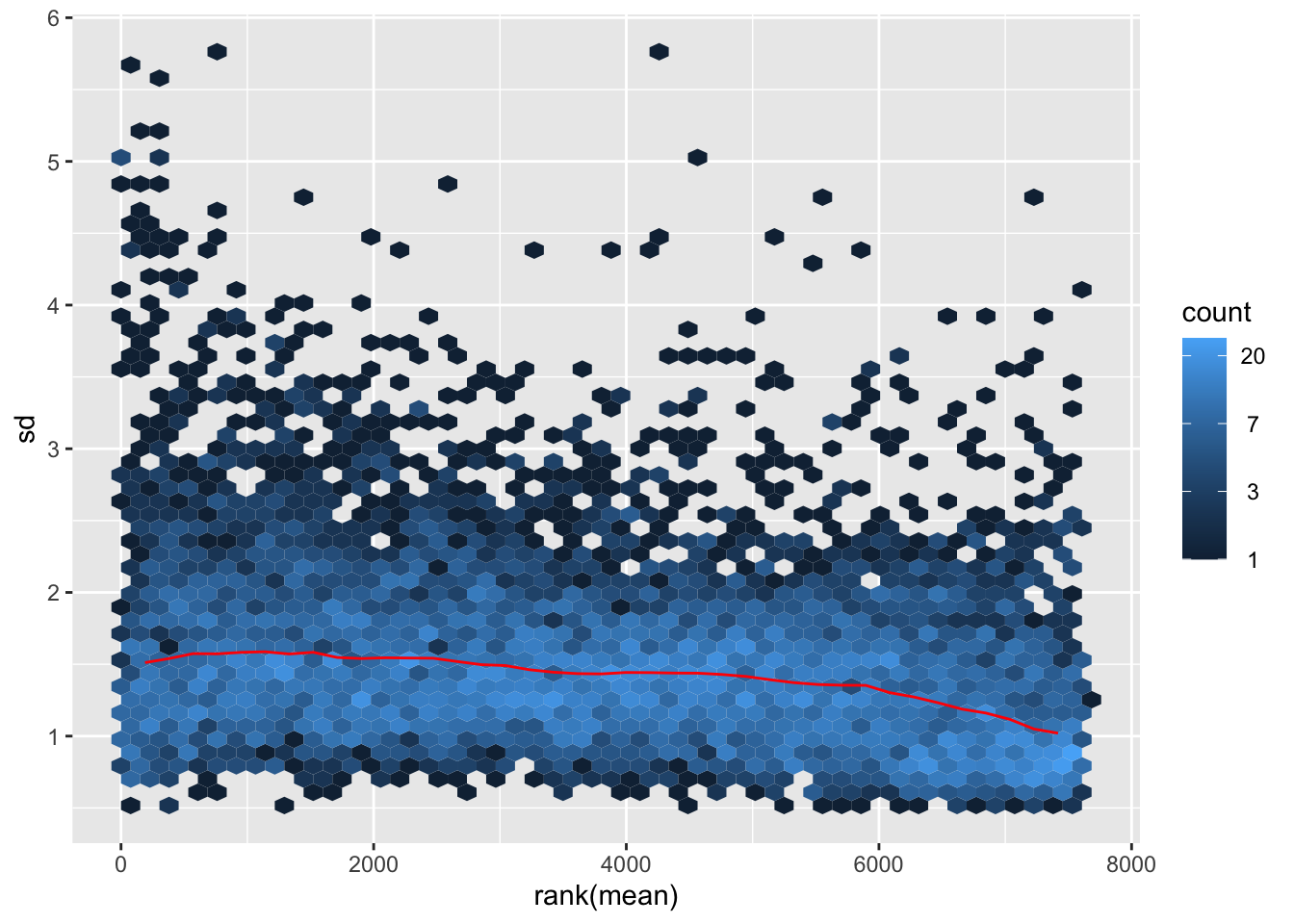 #### Use precursor
#### Use precursor
vsn::meanSdPlot(assay(ppe.pre))
Distribution after normalization
VST
countMat <- assay(ppe.vst)
annoTab <- colData(ppe.vst)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
log2+median
countMat <- assay(ppe.log2Med)
annoTab <- colData(ppe.log2Med)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))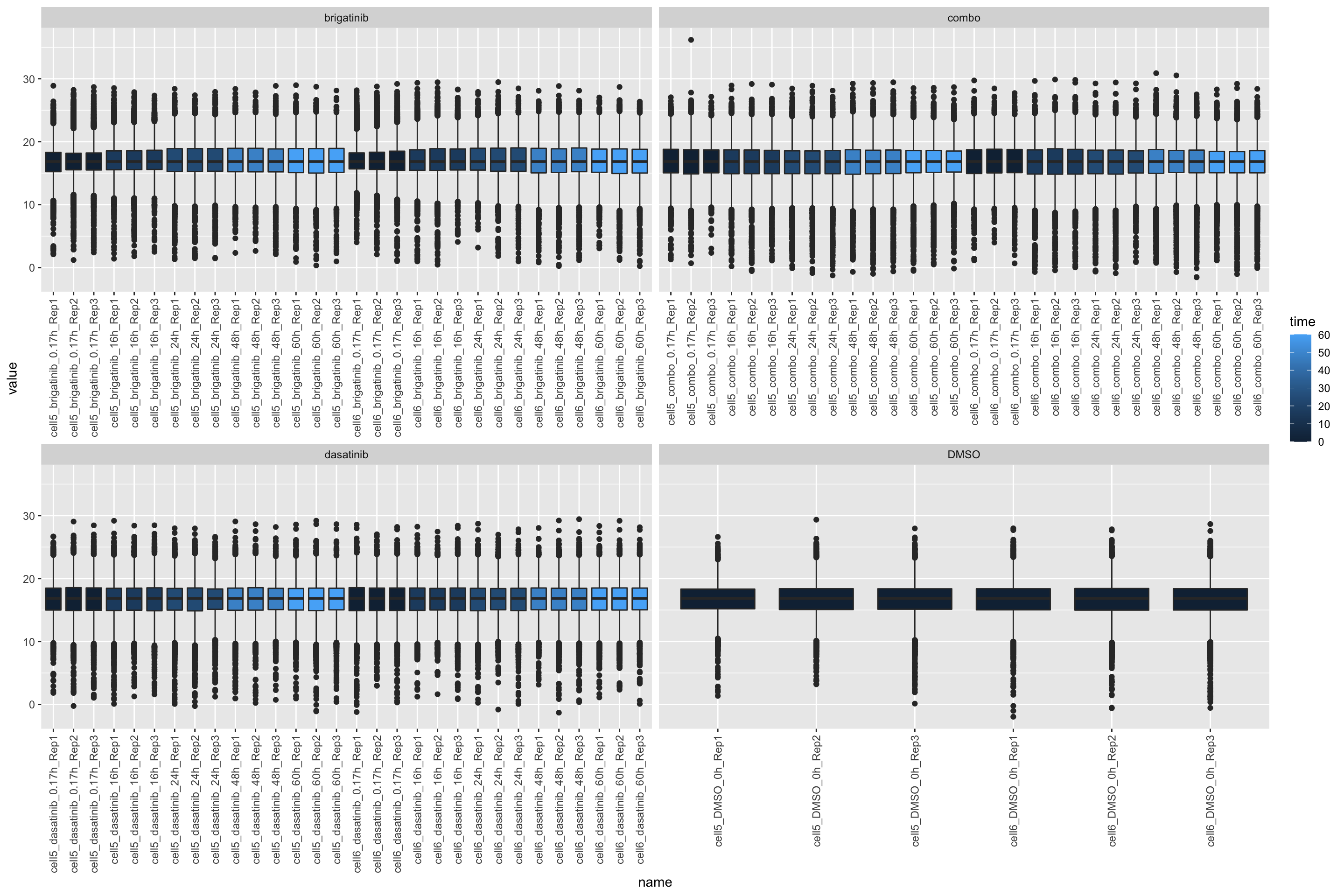 #### only transformation
#### only transformation
countMat <- assay(ppe.log2Only)
annoTab <- colData(ppe.log2Only)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
use precursor
countMat <- assay(ppe.pre)
annoTab <- colData(ppe.pre)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
PCA
VST
exprMat <- assays(ppe.vst)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.vst) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2+median
exprMat <- assays(ppe.log2Med)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Med) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
log2 only
exprMat <- assays(ppe.log2Only)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.log2Only) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Use precursor
exprMat <- assays(ppe.pre)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:2000],]
smpAnno <- colData(ppe.pre) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
VST
colAnno <- colData(ppe.vst)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2+median
colAnno <- colData(ppe.log2Med)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
log2 only
colAnno <- colData(ppe.log2Only)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
precursor
colAnno <- colData(ppe.pre)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Compare the reproducibiliy of replicates when using different normalization methods
sdTab.vst <- getSdTab(ppe.vst) %>% mutate(norm = "vst")
sdTab.log2Med <- getSdTab(ppe.log2Med) %>% mutate(norm = "log2Med")
sdTab.log2Only <- getSdTab(ppe.log2Only) %>% mutate(norm = "log2Only")
sdTab.pre <- getSdTab(ppe.pre) %>% mutate(norm = "precursor")
sumTab <- bind_rows(sdTab.vst, sdTab.log2Med, sdTab.log2Only, sdTab.pre) Add annotations
colTab <- colData(ppe.vst) %>% as_tibble() %>%
distinct(sampleCondi,.keep_all = TRUE)
sumTab <- left_join(sumTab, colTab)Overall distribution
ggplot(sumTab, aes(x=sdVal, fill = norm)) +
geom_histogram(position = "identity", alpha=0.5, color = "grey50") +
xlim(0,6)
Per sample
ggplot(sumTab, aes(x=sampleCondi, y = sdVal, fill = norm)) +
geom_violin() +
facet_wrap(~drug, scale="free_x")+
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
Mean versus SD
ggplot(sumTab, aes(x=meanVal, y=sdVal)) +
geom_hex() + geom_smooth() +
facet_wrap(~norm, ncol=1)
Rank of sd for each normalization method
ordTab <- group_by(sumTab, sampleCondi, rowID) %>%
mutate(index = order(sdVal))
ggplot(ordTab, aes(x=index, fill = norm)) +
geom_bar(position = "dodge", alpha=0.5, color = "grey50") 
For each sample, which normalization method gives the best reproduciblity for replicates
ordPerTab <- arrange(ordTab, index) %>% distinct(sampleCondi, drug,.keep_all = TRUE) %>%
group_by(sampleCondi, norm, drug) %>% summarise(n=length(rowID))
ggplot(ordPerTab, aes(x=sampleCondi, y = n, fill = norm)) +
geom_bar(stat="identity")+
facet_wrap(~drug, scale = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))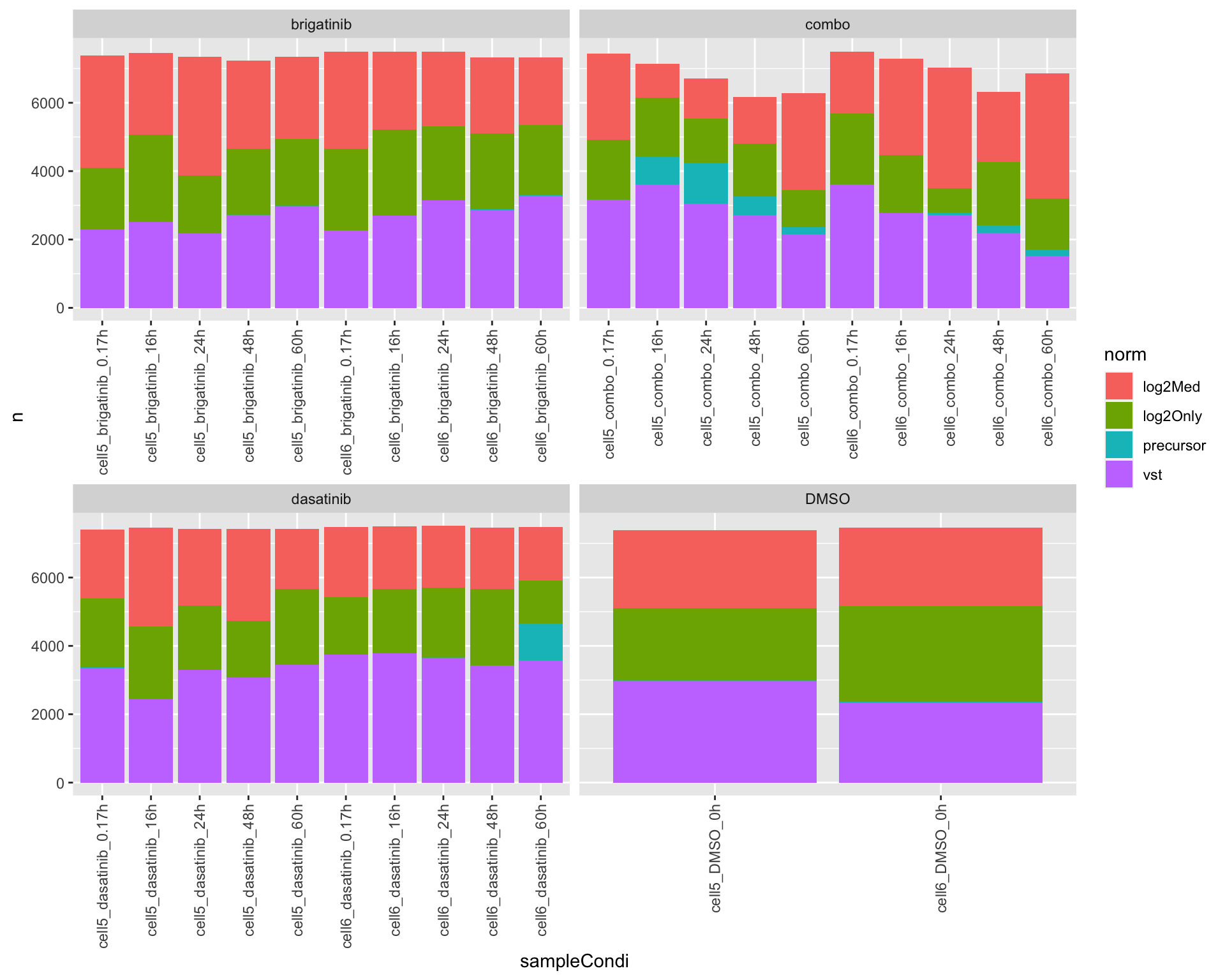 It seems log2 + median scaling can get the best reproducibility among
replicates. No normalization is the worst.
It seems log2 + median scaling can get the best reproducibility among
replicates. No normalization is the worst.
Phosphoproteomic with proteomic regressed out
Preprocessing
ppeSub <- preprocessPhos(maeData, normalize = TRUE, transform = "none", assayName = "PhosReg")[1] "Number of proteins and samples:"
[1] 3574 96Distribution after normalization
countMat <- assay(ppeSub)
annoTab <- colData(ppeSub)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
PCA
exprMat <- assays(ppeSub)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:1000],]
smpAnno <- colData(ppeSub) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
colAnno <- colData(ppeSub)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Remove one potential outlier
ppeSub <- ppeSub[, colnames(ppeSub)!="cell5_combo_24h_Rep2"]Redo PCA
exprMat <- assays(ppeSub)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:1000],]
smpAnno <- colData(ppeSub) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
colAnno <- colData(ppeSub)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Use ration between phosphoproteome and proteome
Preprocessing
ppeSub <- preprocessPhos(maeData, normalize = TRUE, transform = "none", assayName = "PhosRatio")[1] "Number of proteins and samples:"
[1] 3574 96Distribution after normalization
countMat <- assay(ppeSub)
annoTab <- colData(ppeSub)[,c("sample","time","drug")] %>% as_tibble()
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
left_join(annoTab, by = c(name = "sample"))ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot(aes(fill = time)) +
facet_wrap(~drug, scales = "free_x") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
PCA
exprMat <- assays(ppeSub)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:1000],]
smpAnno <- colData(ppeSub) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
colAnno <- colData(ppeSub)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
Remove one potential outlier
ppeSub <- ppeSub[, colnames(ppeSub)!="cell5_combo_24h_Rep2"]Redo PCA
exprMat <- assays(ppeSub)[["imputed"]]
sds <- genefilter::rowSds(exprMat)
exprMat <- exprMat[order(sds, decreasing = TRUE)[1:1000],]
smpAnno <- colData(ppeSub) %>%
as_tibble(rownames = "id")
pcRes <- prcomp(t(exprMat), scale. = TRUE, center = TRUE)
pcTab <- pcRes$x[,1:10] %>%
as_tibble(rownames = "id") %>%
left_join(smpAnno)PC1 versus PC2
ggplot(pcTab, aes(x=PC1, y=PC2)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
PC3 versus PC4
ggplot(pcTab, aes(x=PC3, y=PC4)) +
geom_point(aes(col = drug, size = factor(time),
shape = replicate)) +
ggrepel::geom_text_repel(aes(label = id)) +
theme_bw()
Hierarchical clustering
colAnno <- colData(ppeSub)[,c("cellLine","drug","time")] %>% data.frame()
exprMat.scaled <- jyluMisc::mscale(exprMat, center = TRUE, scale = TRUE, censor = 5)
pheatmap::pheatmap(exprMat.scaled, annotation_col = colAnno, clustering_method = "ward.D2",
color = colorRampPalette(c("blue","white","red"))(100),
breaks = seq(-5,5, length.out = 101),
show_rownames = FALSE)
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] forcats_0.5.1 stringr_1.4.1
[3] dplyr_1.0.9 purrr_0.3.4
[5] readr_2.1.2 tidyr_1.2.0
[7] tibble_3.1.8 ggplot2_3.3.6
[9] tidyverse_1.3.2 proDA_1.10.0
[11] MultiAssayExperiment_1.22.0 SummarizedExperiment_1.26.1
[13] Biobase_2.56.0 GenomicRanges_1.48.0
[15] GenomeInfoDb_1.32.2 IRanges_2.30.0
[17] S4Vectors_0.34.0 BiocGenerics_0.42.0
[19] MatrixGenerics_1.8.1 matrixStats_0.62.0
loaded via a namespace (and not attached):
[1] GGally_2.1.2 exactRankTests_0.8-35 coda_0.19-4
[4] bit64_4.0.5 knitr_1.39 multcomp_1.4-19
[7] DelayedArray_0.22.0 data.table_1.14.2 KEGGREST_1.36.3
[10] RCurl_1.98-1.7 doParallel_1.0.17 generics_0.1.3
[13] preprocessCore_1.58.0 cowplot_1.1.1 TH.data_1.1-1
[16] RSQLite_2.2.15 proxy_0.4-27 bit_4.0.4
[19] tzdb_0.3.0 xml2_1.3.3 lubridate_1.8.0
[22] httpuv_1.6.6 assertthat_0.2.1 viridis_0.6.2
[25] gargle_1.2.0 xfun_0.31 hms_1.1.1
[28] jquerylib_0.1.4 evaluate_0.15 promises_1.2.0.1
[31] fansi_1.0.3 caTools_1.18.2 dendextend_1.16.0
[34] dbplyr_2.2.1 readxl_1.4.0 km.ci_0.5-6
[37] igraph_1.3.4 DBI_1.1.3 htmlwidgets_1.5.4
[40] reshape_0.8.9 googledrive_2.0.0 ellipsis_0.3.2
[43] jyluMisc_0.1.5 ggpubr_0.4.0 backports_1.4.1
[46] annotate_1.74.0 PhosR_1.6.0 vctrs_0.4.1
[49] imputeLCMD_2.1 abind_1.4-5 cachem_1.0.6
[52] withr_2.5.0 cluster_2.1.3 crayon_1.5.2
[55] drc_3.0-1 relations_0.6-12 genefilter_1.78.0
[58] pkgconfig_2.0.3 slam_0.1-50 labeling_0.4.2
[61] nlme_3.1-158 ProtGenerics_1.28.0 rlang_1.0.6
[64] lifecycle_1.0.3 sandwich_3.0-2 affyio_1.66.0
[67] modelr_0.1.8 cellranger_1.1.0 rprojroot_2.0.3
[70] Matrix_1.4-1 KMsurv_0.1-5 carData_3.0-5
[73] zoo_1.8-10 DEP_1.18.0 reprex_2.0.1
[76] GlobalOptions_0.1.2 googlesheets4_1.0.0 pheatmap_1.0.12
[79] png_0.1-7 viridisLite_0.4.0 rjson_0.2.21
[82] mzR_2.30.0 bitops_1.0-7 shinydashboard_0.7.2
[85] visNetwork_2.1.0 KernSmooth_2.23-20 Biostrings_2.64.0
[88] blob_1.2.3 workflowr_1.7.0 shape_1.4.6
[91] maxstat_0.7-25 rstatix_0.7.0 tmvtnorm_1.5
[94] ggsignif_0.6.3 scales_1.2.0 memoise_2.0.1
[97] magrittr_2.0.3 plyr_1.8.7 hexbin_1.28.2
[100] gplots_3.1.3 zlibbioc_1.42.0 compiler_4.2.0
[103] RColorBrewer_1.1-3 plotrix_3.8-2 pcaMethods_1.88.0
[106] clue_0.3-61 cli_3.4.1 affy_1.74.0
[109] XVector_0.36.0 mgcv_1.8-40 MASS_7.3-58
[112] tidyselect_1.1.2 vsn_3.64.0 stringi_1.7.8
[115] highr_0.9 yaml_2.3.5 norm_1.0-10.0
[118] MALDIquant_1.21 ggrepel_0.9.1 survMisc_0.5.6
[121] grid_4.2.0 sass_0.4.2 fastmatch_1.1-3
[124] tools_4.2.0 ruv_0.9.7.1 parallel_4.2.0
[127] circlize_0.4.15 rstudioapi_0.13 MsCoreUtils_1.8.0
[130] foreach_1.5.2 git2r_0.30.1 gridExtra_2.3
[133] farver_2.1.1 mzID_1.34.0 digest_0.6.30
[136] BiocManager_1.30.18 shiny_1.7.3 Rcpp_1.0.9
[139] car_3.1-0 broom_1.0.0 later_1.3.0
[142] ncdf4_1.19 survminer_0.4.9 httr_1.4.3
[145] MSnbase_2.22.0 ggdendro_0.1.23 AnnotationDbi_1.58.0
[148] ComplexHeatmap_2.12.0 colorspace_2.0-3 rvest_1.0.2
[151] XML_3.99-0.10 fs_1.5.2 splines_4.2.0
[154] gmm_1.6-6 xtable_1.8-4 jsonlite_1.8.3
[157] marray_1.74.0 R6_2.5.1 sets_1.0-21
[160] pillar_1.8.0 htmltools_0.5.3 mime_0.12
[163] glue_1.6.2 fastmap_1.1.0 DT_0.23
[166] BiocParallel_1.30.3 class_7.3-20 codetools_0.2-18
[169] fgsea_1.22.0 mvtnorm_1.1-3 utf8_1.2.2
[172] lattice_0.20-45 bslib_0.4.1 network_1.17.2
[175] gtools_3.9.3 shinyjs_2.1.0 survival_3.4-0
[178] limma_3.52.2 rmarkdown_2.14 statnet.common_4.6.0
[181] munsell_0.5.0 e1071_1.7-11 GetoptLong_1.0.5
[184] GenomeInfoDbData_1.2.8 iterators_1.0.14 piano_2.12.0
[187] impute_1.70.0 haven_2.5.0 reshape2_1.4.4
[190] gtable_0.3.0