Analysis of differentially expressed proteins/RNAs related to trisomy19 and trisomy12
Junyan Lu
2020-03-13
Last updated: 2020-04-25
Checks: 6 1
Knit directory: Proteomics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200227) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/correlateGenomic_PC12adjusted_cache/
Ignored: analysis/correlateGenomic_cache/
Ignored: analysis/correlateGenomic_noBlock_MCLL_cache/
Ignored: analysis/correlateGenomic_noBlock_UCLL_cache/
Ignored: analysis/correlateGenomic_noBlock_cache/
Ignored: code/.Rhistory
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/analysisSplicing.Rmd
Untracked: analysis/analysisTrisomy19.Rmd
Untracked: analysis/annotateCNV.Rmd
Untracked: analysis/correlateGenomic_PC12adjusted.Rmd
Untracked: analysis/correlateGenomic_noBlock.Rmd
Untracked: analysis/correlateGenomic_noBlock_MCLL.Rmd
Untracked: analysis/correlateGenomic_noBlock_UCLL.Rmd
Untracked: analysis/default.css
Untracked: analysis/patCNV.csv
Untracked: analysis/patCNV.xlsx
Untracked: analysis/patSNV.csv
Untracked: analysis/peptideValidate.Rmd
Untracked: analysis/processPeptides_LUMOS.Rmd
Untracked: analysis/style.css
Untracked: code/utils.R
Untracked: data/190909_CLL_prot_abund_med_norm.tsv
Untracked: data/190909_CLL_prot_abund_no_norm.tsv
Untracked: data/20190423_Proteom_submitted_samples_bereinigt.xlsx
Untracked: data/20191025_Proteom_submitted_samples_final.xlsx
Untracked: data/LUMOS/
Untracked: data/LUMOS_peptides/
Untracked: data/LUMOS_protAnnotation.csv
Untracked: data/LUMOS_protAnnotation_fix.csv
Untracked: data/SampleAnnotation_cleaned.xlsx
Untracked: data/facTab_IC50atLeast3New.RData
Untracked: data/gmts/
Untracked: data/mapEnsemble.txt
Untracked: data/mapSymbol.txt
Untracked: data/pyprophet_export_aligned.csv
Untracked: data/timsTOF_protAnnotation.csv
Untracked: output/LUMOS_processed.RData
Untracked: output/dxdCLL.RData
Untracked: output/pepCLL_lumos.RData
Untracked: output/pepTab_lumos.RData
Untracked: output/proteomic_LUMOS_20200227.RData
Untracked: output/proteomic_LUMOS_20200320.RData
Untracked: output/proteomic_timsTOF_20200227.RData
Untracked: output/splicingResults.RData
Untracked: output/timsTOF_processed.RData
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/analysisSF3B1.Rmd
Modified: analysis/compareProteomicsRNAseq.Rmd
Modified: analysis/correlateGenomic.Rmd
Deleted: analysis/correlateGenomic_removePC.Rmd
Modified: analysis/correlateMIR.Rmd
Modified: analysis/correlateMethylationCluster.Rmd
Modified: analysis/index.Rmd
Modified: analysis/predictOutcome.Rmd
Modified: analysis/processProteomics_LUMOS.Rmd
Modified: analysis/qualityControl_LUMOS.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Proteomics data
Background of trisomy19 patients in the complete CLL cohort
tri19Tab <- filter(patMeta, diagnosis %in% "CLL") %>%
select(Patient.ID, IGHV.status, trisomy12, trisomy19) %>%
filter(trisomy19 %in% 1)
tri19Tab# A tibble: 7 x 4
Patient.ID IGHV.status trisomy12 trisomy19
<chr> <fct> <fct> <fct>
1 P0045 M 1 1
2 P0098 M 1 1
3 P0494 M 1 1
4 P0519 M 1 1
5 P0615 M 1 1
6 P0622 U 0 1
7 P0648 M 1 1 There are now 7 patients with trisomy19 in our cohort, most of them are M-CLL patients with trisomy12 One U-CLL patient, P0622, does not have trisomy12 but has trisomy19. This patients does not have FISH, WGS or WES data. The CNV status was inferred from methylation data
Transcriptomic analysis
Preprocessing data
Sample subsetting
Subset sample: select M-CLLs with annotation for trisomy12 and trisomy19
dds$IGHV <- patMeta[match(colnames(dds),patMeta$Patient.ID),]$IGHV.status
dds$trisomy12 <- patMeta[match(colnames(dds),patMeta$Patient.ID),]$trisomy12
dds$trisomy19 <- patMeta[match(colnames(dds),patMeta$Patient.ID),]$trisomy19
ddsSub <- dds[,dds$diag == "CLL" & dds$IGHV %in% "M" & !is.na(dds$trisomy12) & !is.na(dds$trisomy19)]Summary of trisomy12 and trisomy19 status
annoTab <- tibble(patID = colnames(ddsSub),
tri12 = ifelse(ddsSub$trisomy12 == 1, "tri12","wt"),
tri19 = ifelse(ddsSub$trisomy19 ==1, "tri19","wt")) %>%
mutate(group = ifelse(tri12 == "tri12", ifelse(tri19 == "tri19","both","onlyTri12"),"none"))
table(annoTab$tri12, annoTab$tri19)
tri19 wt
tri12 5 7
wt 0 97There are too many samples without either trisomy19 and trisomy12. The sample imbalance will cause bias in hypothesis testing. In addition, the other recurrent alterations in wildtype group may also complicate the problem.
To ensure the compatibility and reduce noise, in the below analysis, I will:
Check the distribution of several recurrent mutations that impact gene expression (based on Almut’s paper), namely: del13q, SF3B1, del17p, TP53, del11q, BRAF, gain8q, del8p, NOTCH1, MED12, ATM.
Get mutations that occurred in non-wildtype samples (with trisomy12, trisomy19 or both).
Remove wild type samples that contain the recurrent mutations that do not occur in non-wildtype samples.
Block for the mutations that occur in both wild type and non-wildtype samples.
#Get the distribution of recurrent mutations in RNAseq samples
patSum <- select(patMeta, Patient.ID, del13q, SF3B1, del17p, TP53, del11q, BRAF, gain8q, del8p, NOTCH1, MED12, ATM) %>%
filter(Patient.ID %in% annoTab$patID) %>%
gather(key = "gene",value = "status",-Patient.ID) %>%
mutate(status = as.integer(status)) %>% filter(status %in% 1)Which mutations occur in non-wildtype samples?
geneKeep <- unique(filter(patSum, Patient.ID %in% filter(annoTab, group != "none")$patID)$gene)
geneKeep[1] "del13q" "SF3B1" "del17p" "TP53" Remove wildtype samples with recurrent mutations except for those four: del13q, SF3B1, del17p and TP53. Those genes will be blocked when doing hypothesis test
patSum.other <- filter(patSum, !gene %in% geneKeep)
annoTab <- filter(annoTab, !(group == "none" & patID %in% patSum.other$Patient.ID))Sample summary after filtering
table(annoTab$tri12, annoTab$tri19)
tri19 wt
tri12 5 7
wt 0 82Filtering transcripts
ddsSub <- ddsSub[,annoTab$patID]
ddsSub$group <- annoTab$group
geneTab <- patMeta[match(ddsSub$PatID, patMeta$Patient.ID),c("Patient.ID", "del13q","SF3B1","del17p","TP53")] %>%
data.frame() %>% column_to_rownames("Patient.ID")
colData(ddsSub) <- cbind(colData(ddsSub),geneTab)
#filter out none protein coding genes and gene on sex chromosome
ddsSub<-ddsSub[rowData(ddsSub)$biotype %in% "protein_coding",]
ddsSub <- ddsSub[! rowData(ddsSub)$symbol %in% c("",NA),]
##vst
ddsSub.vst <- varianceStabilizingTransformation(ddsSub)
dim(ddsSub)[1] 20074 94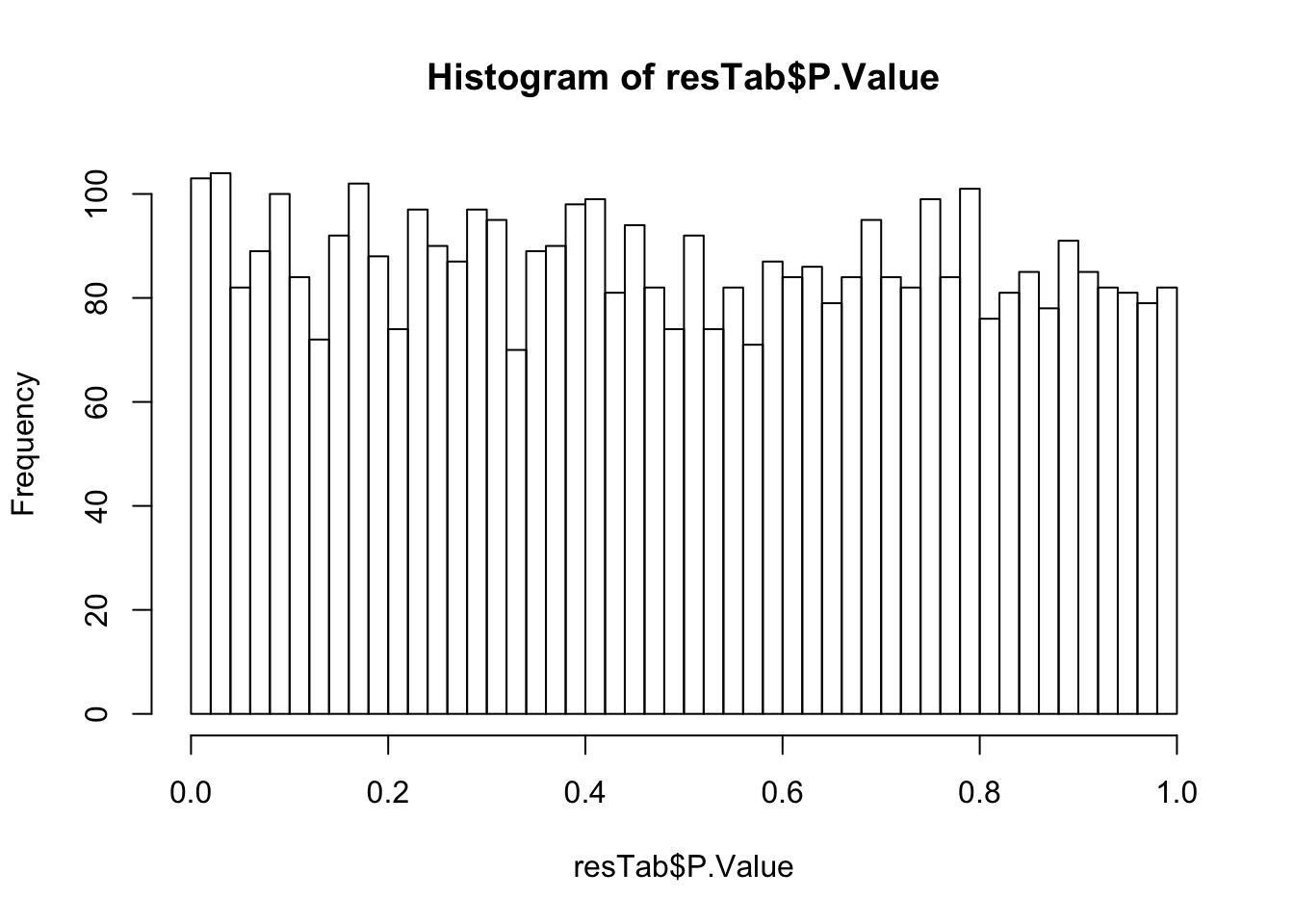
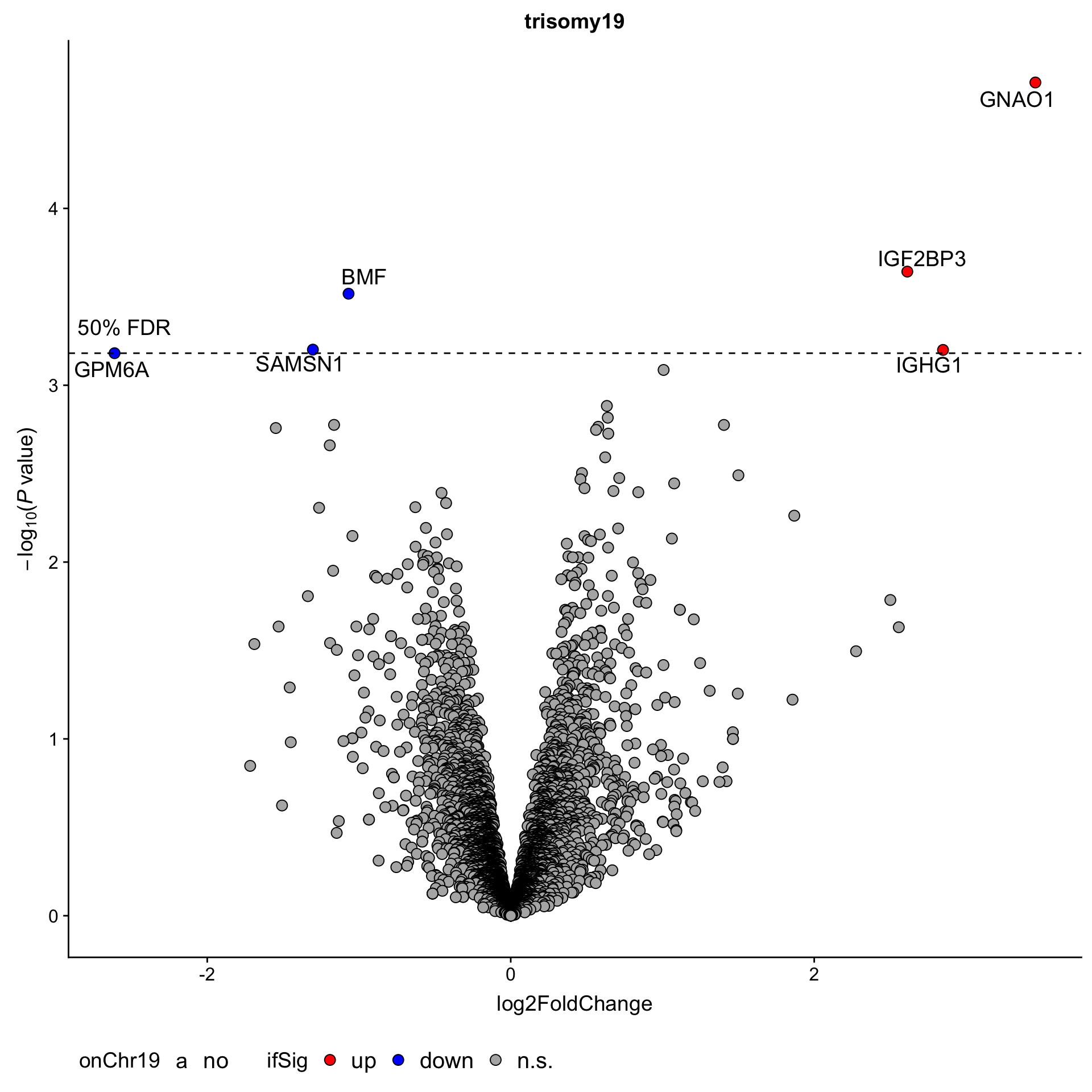
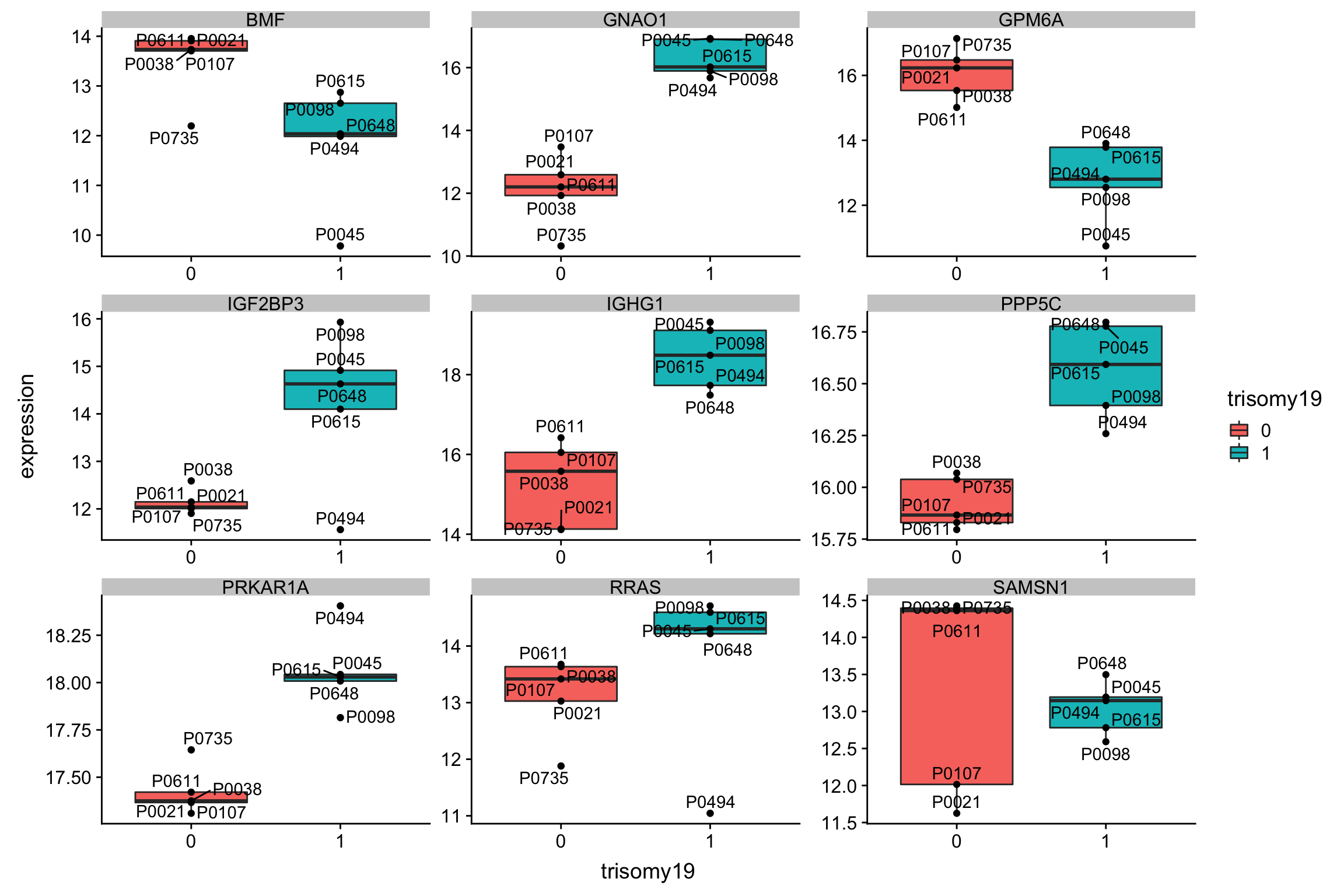 Although the separation looks good, the effect sizes are generally small. This may also lead to high adjusted p-values
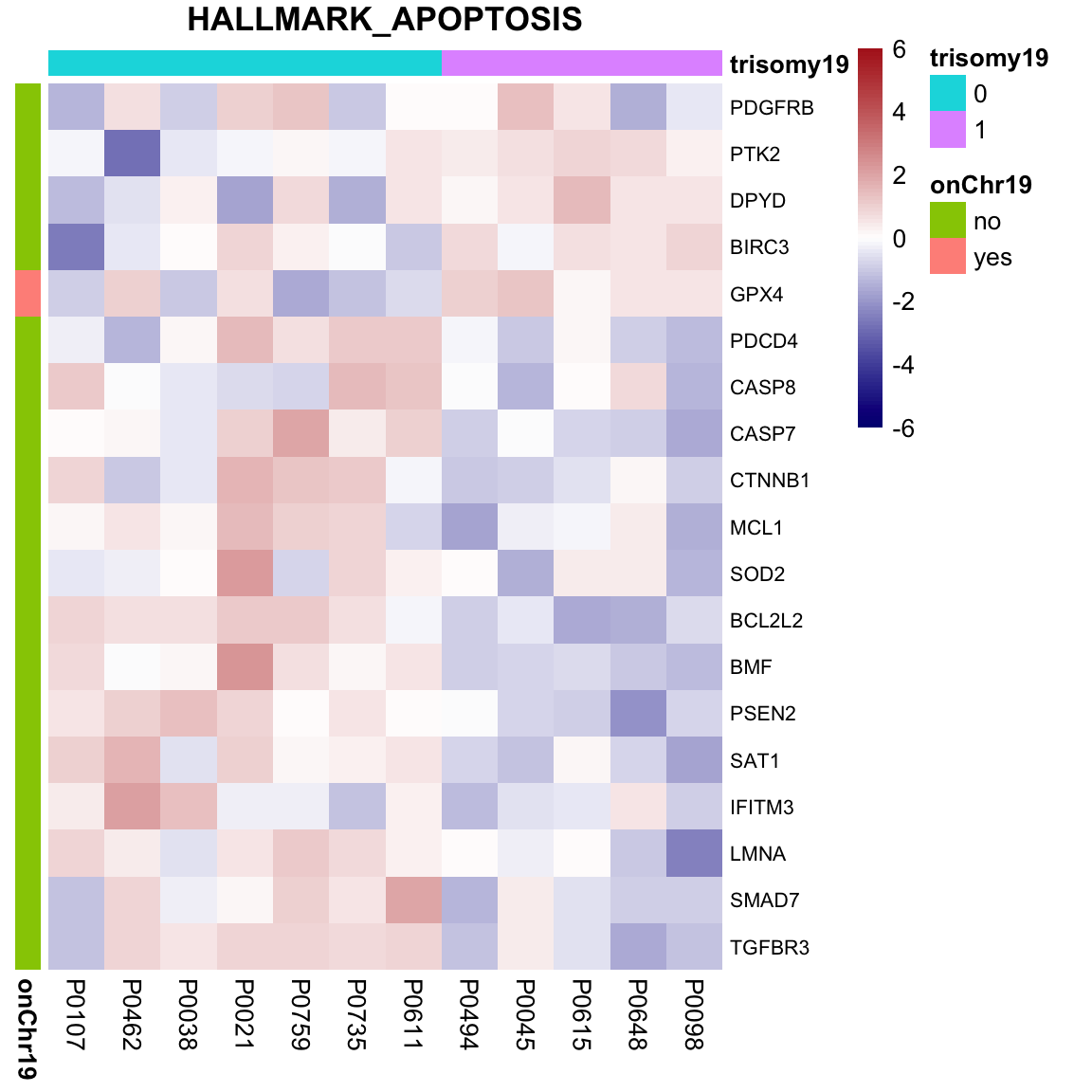
Although the separation looks good, the effect sizes are generally small. This may also lead to high adjusted p-values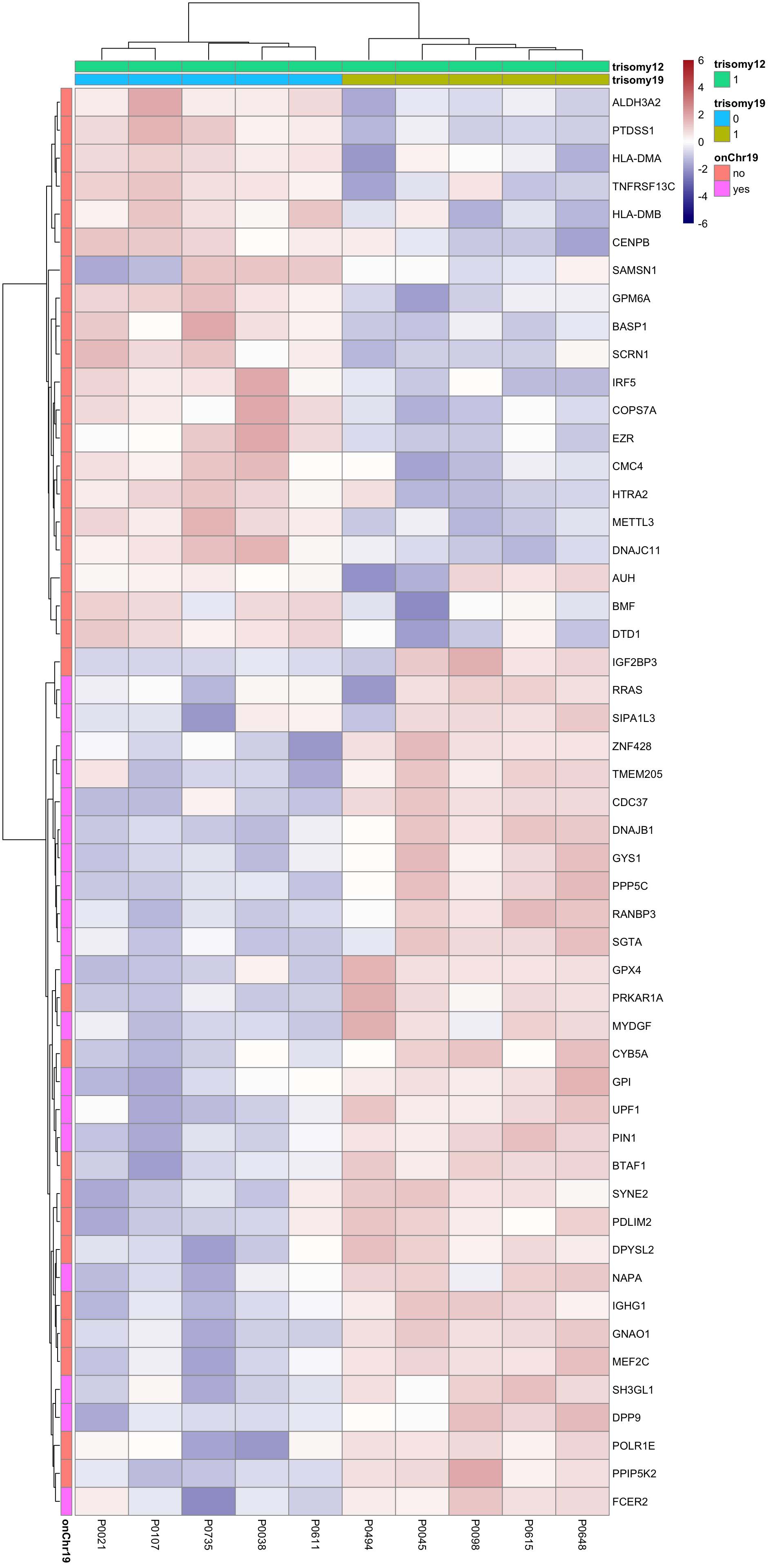 Trisomy19 samples form its own cluster.
Trisomy19 samples form its own cluster.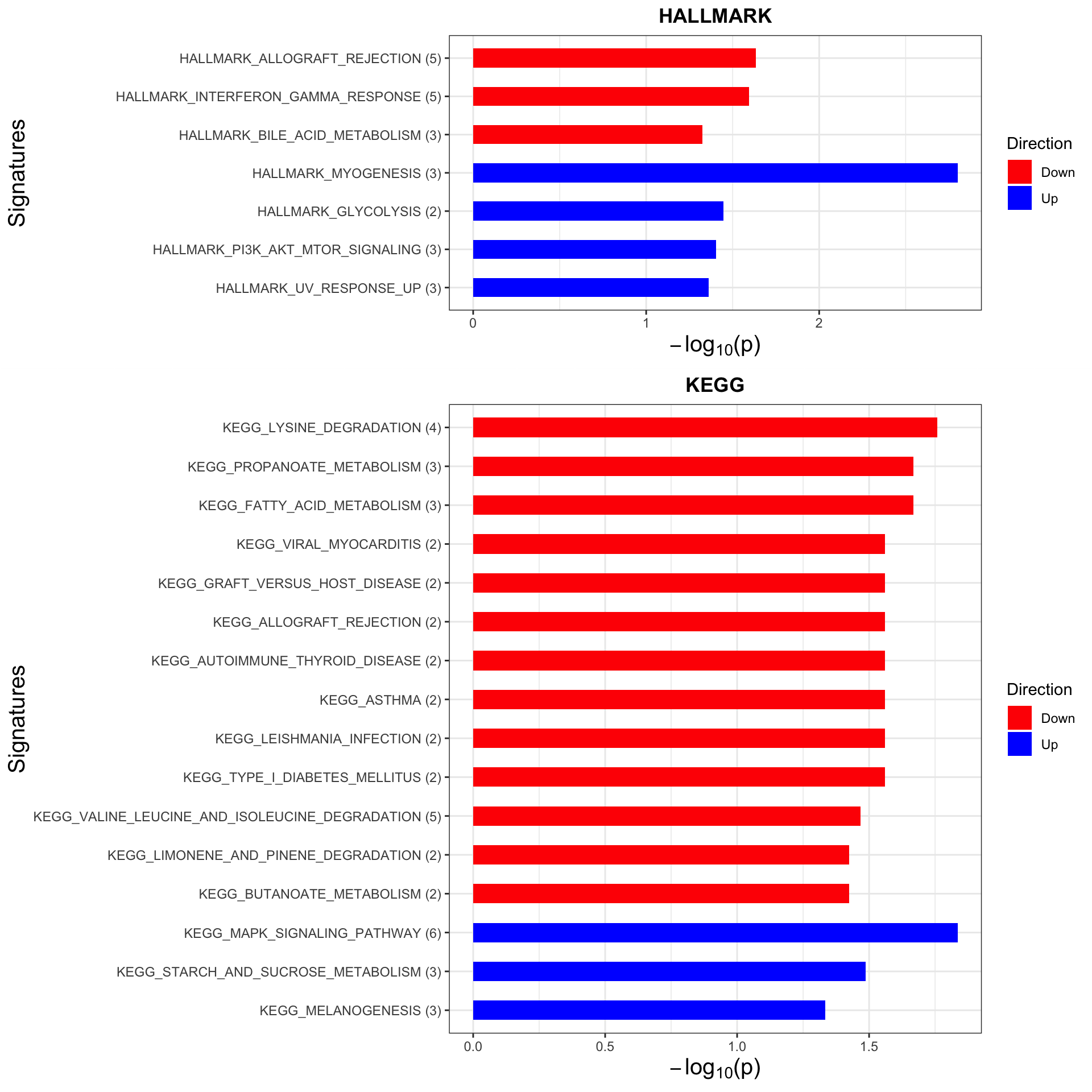
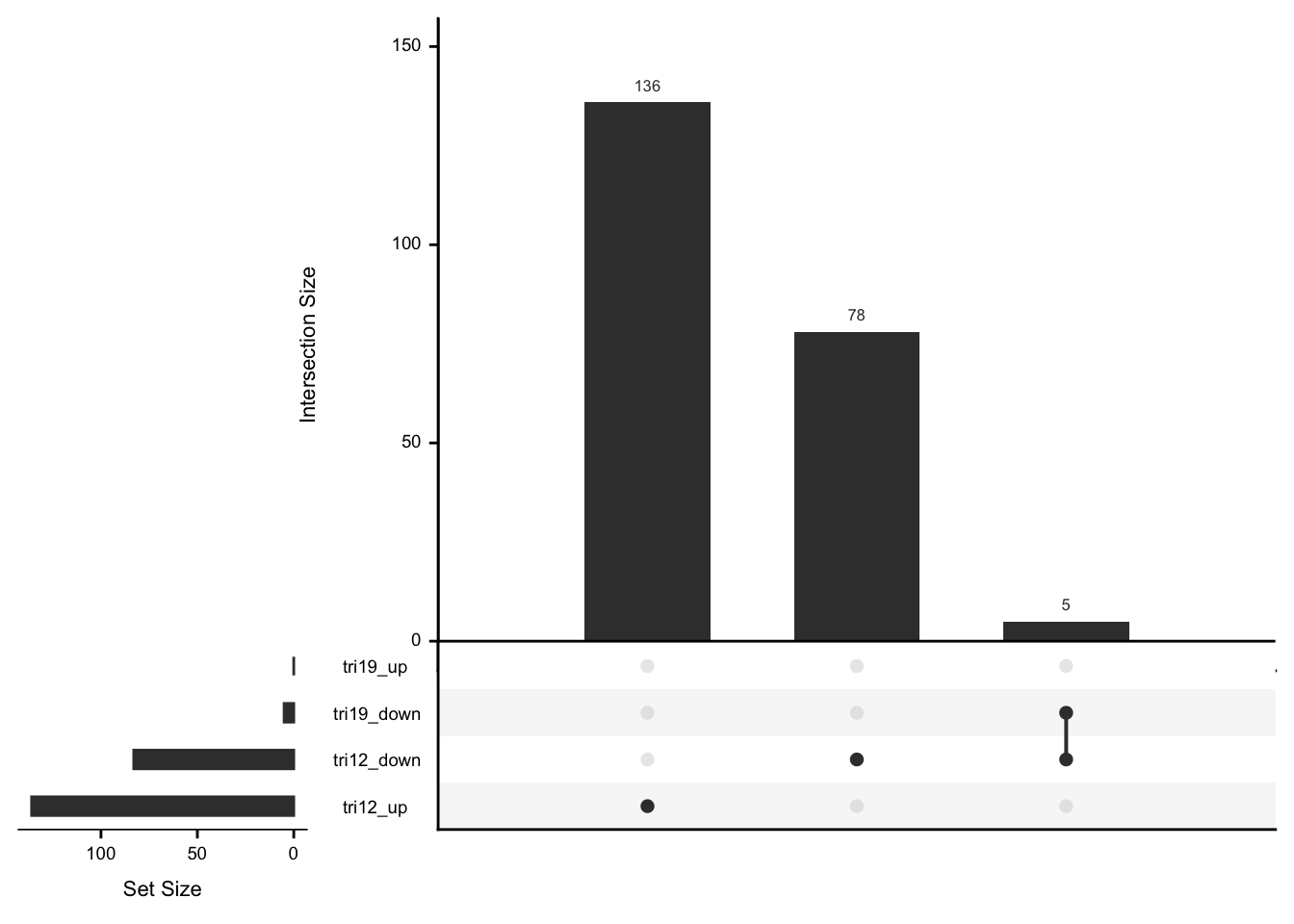
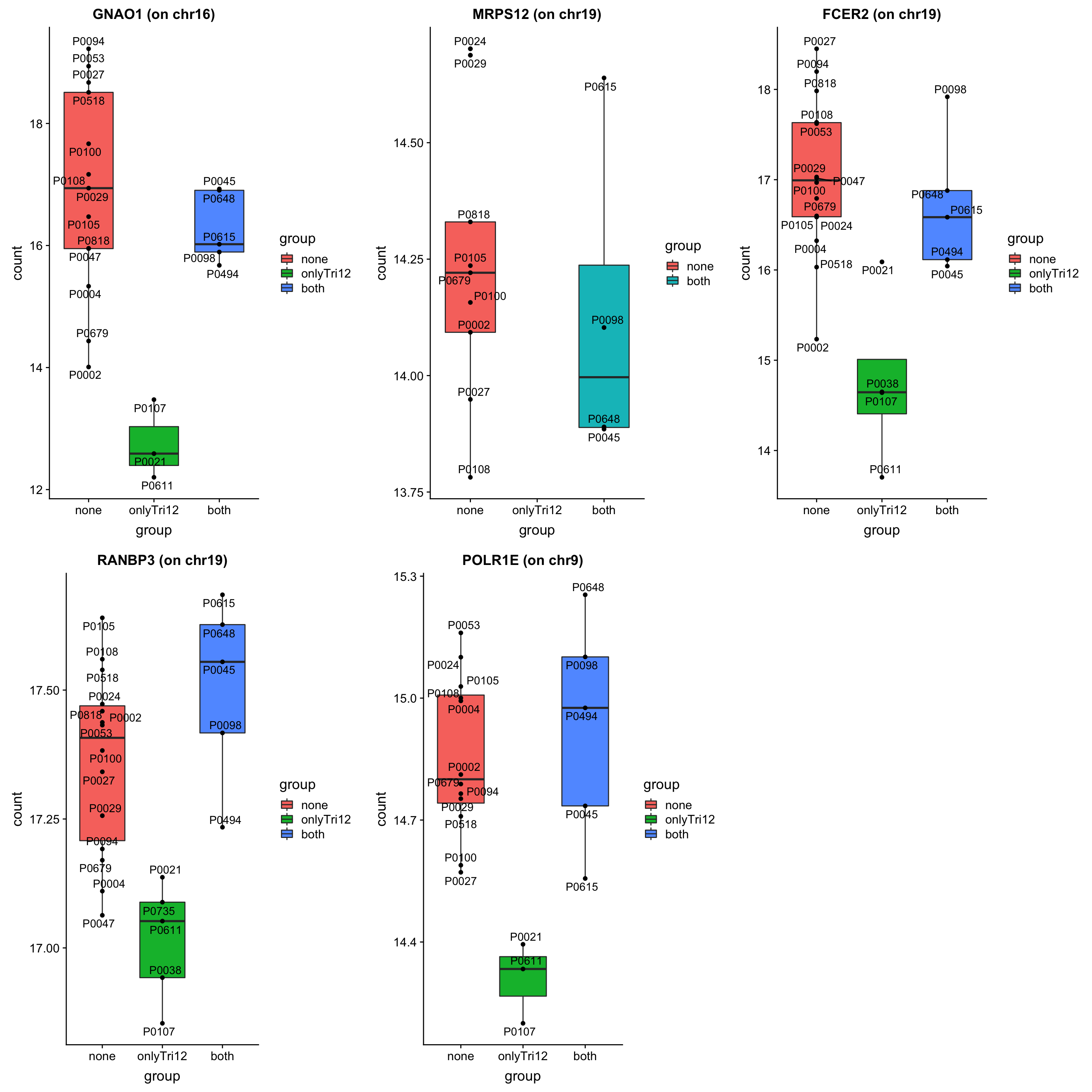 As we don’t have samples with only trisomy19 but not trisomy12, we don’t have a signature of “pure” trisomy19 effect on expression. So those are not “epistatic interactions”. But can be considered as additional effect of trisomy19 on the genes related to trisomy12.
As we don’t have samples with only trisomy19 but not trisomy12, we don’t have a signature of “pure” trisomy19 effect on expression. So those are not “epistatic interactions”. But can be considered as additional effect of trisomy19 on the genes related to trisomy12.
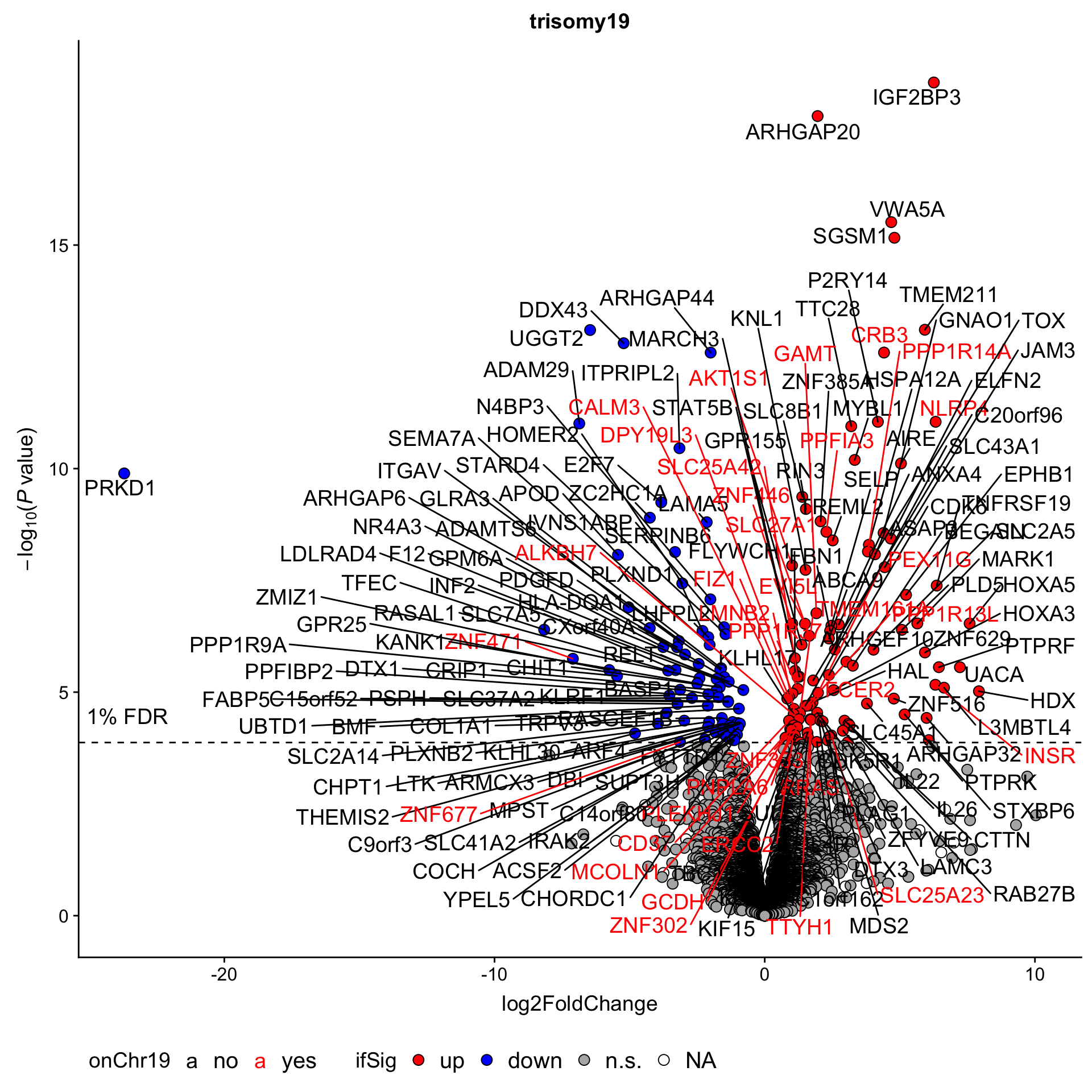
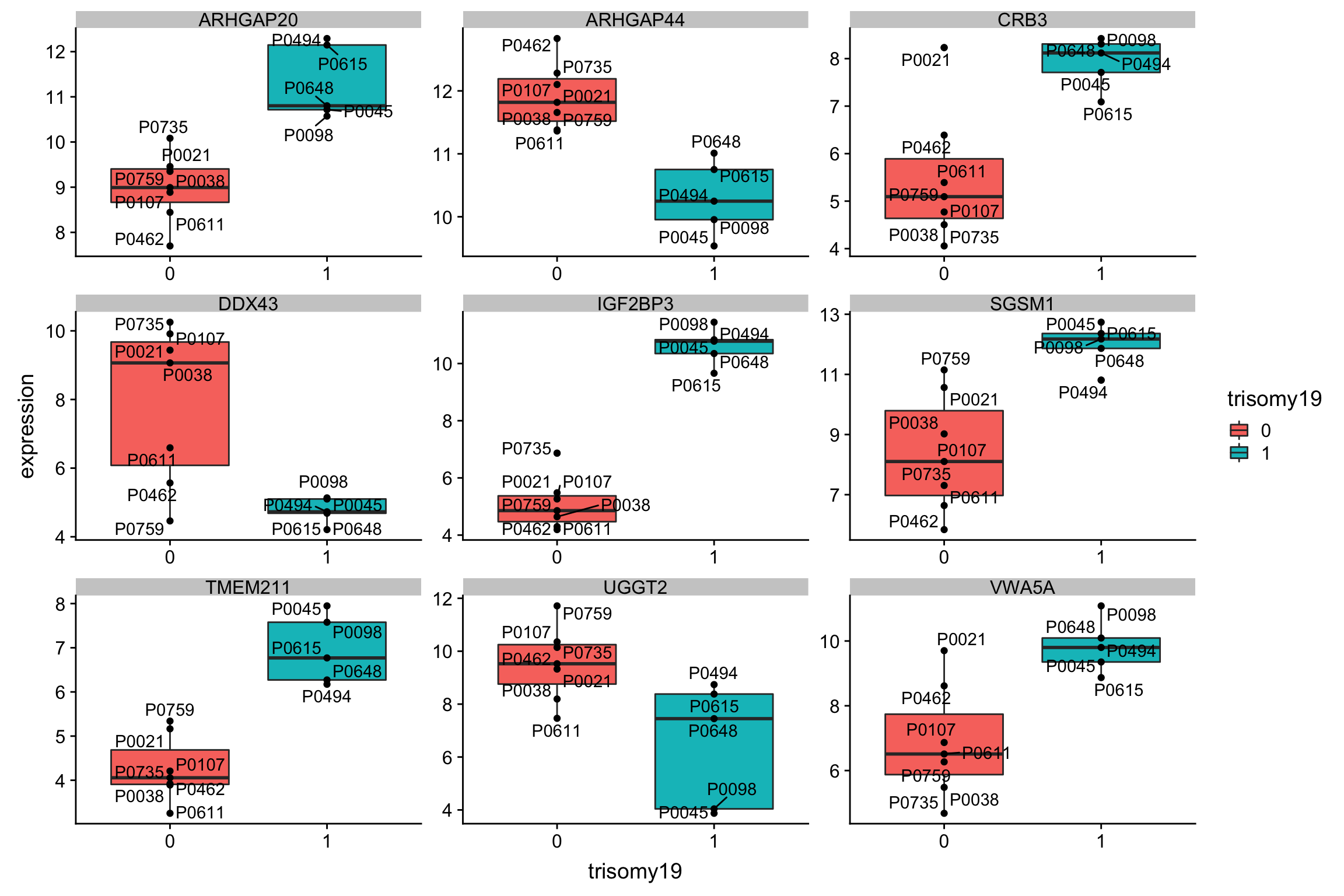
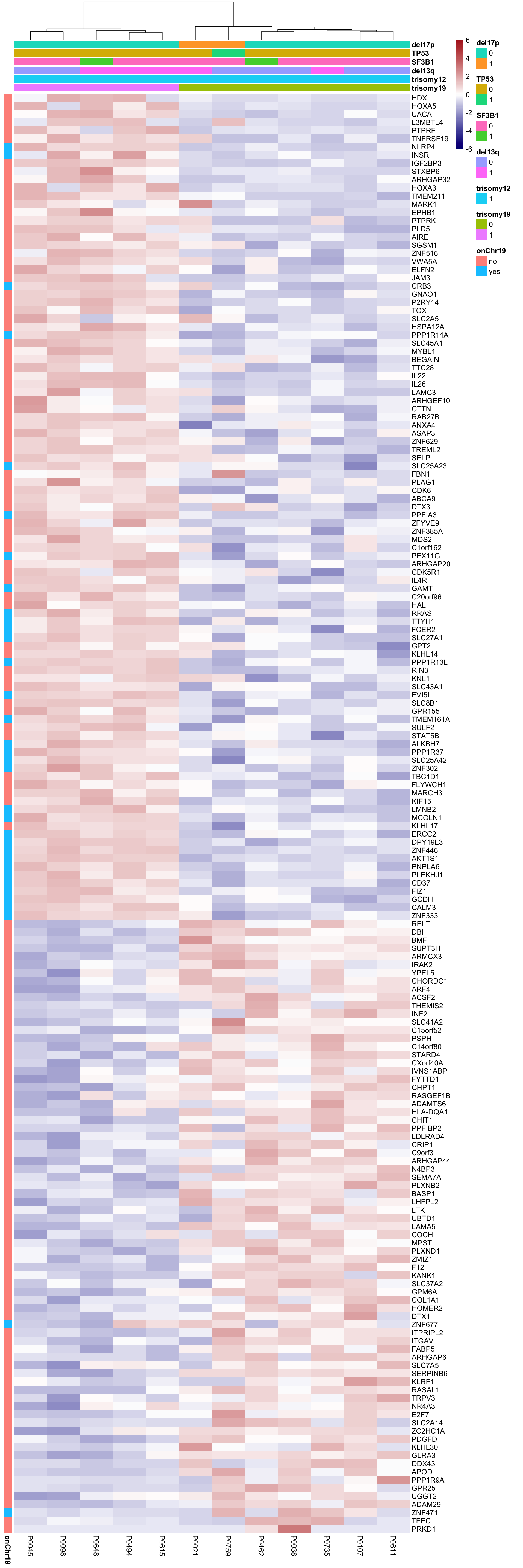 Trisomy19 samples form its own cluster.
Trisomy19 samples form its own cluster.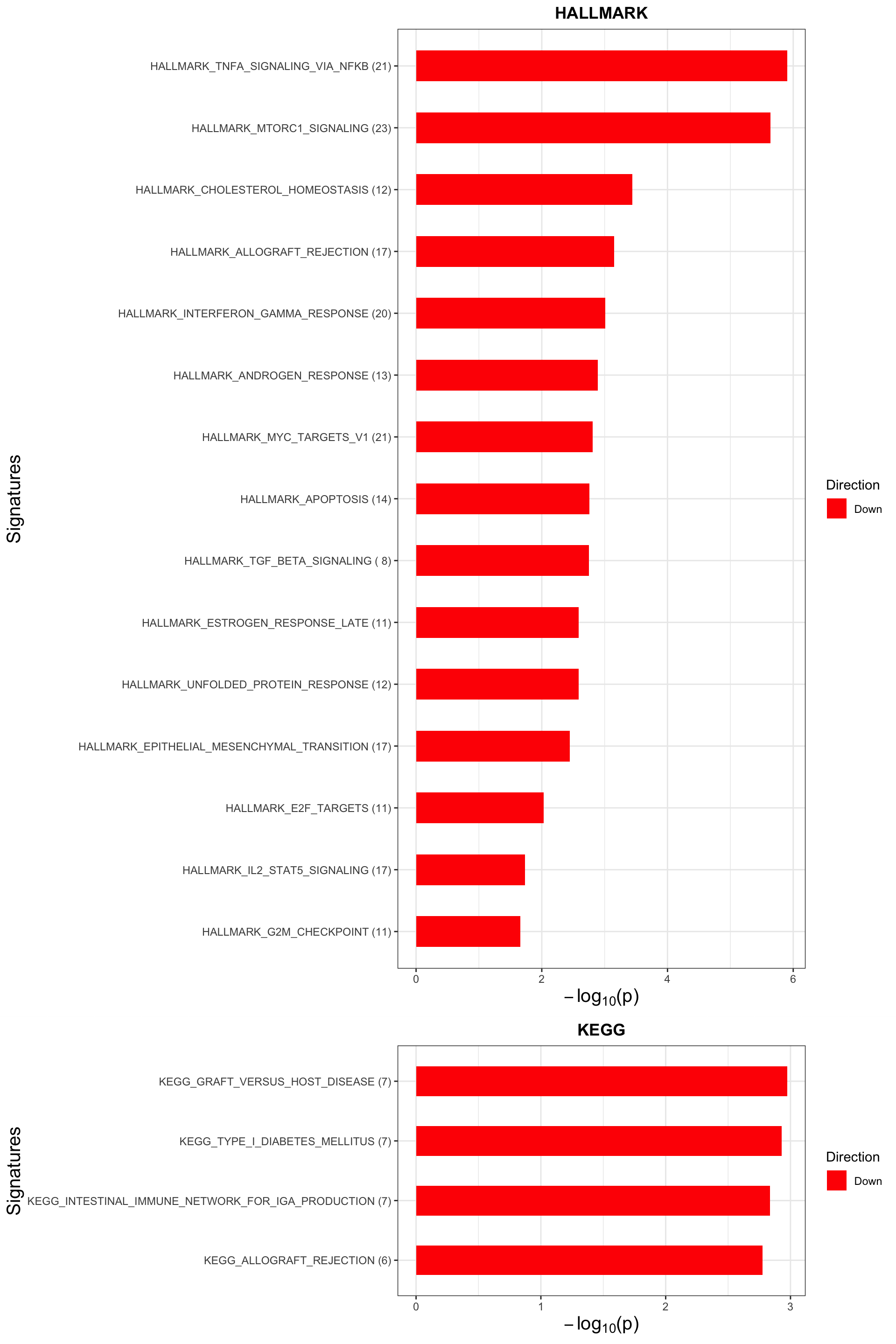
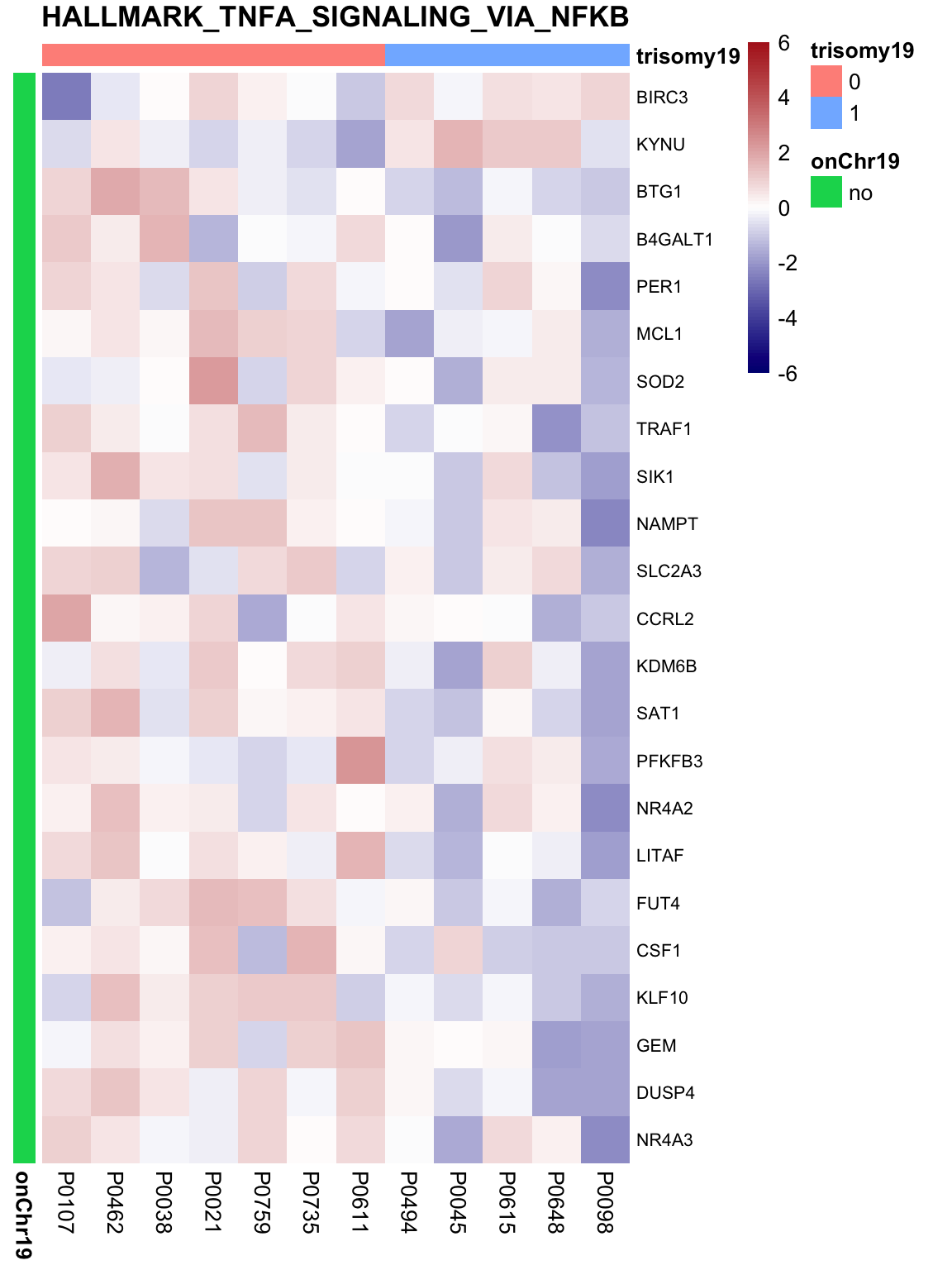
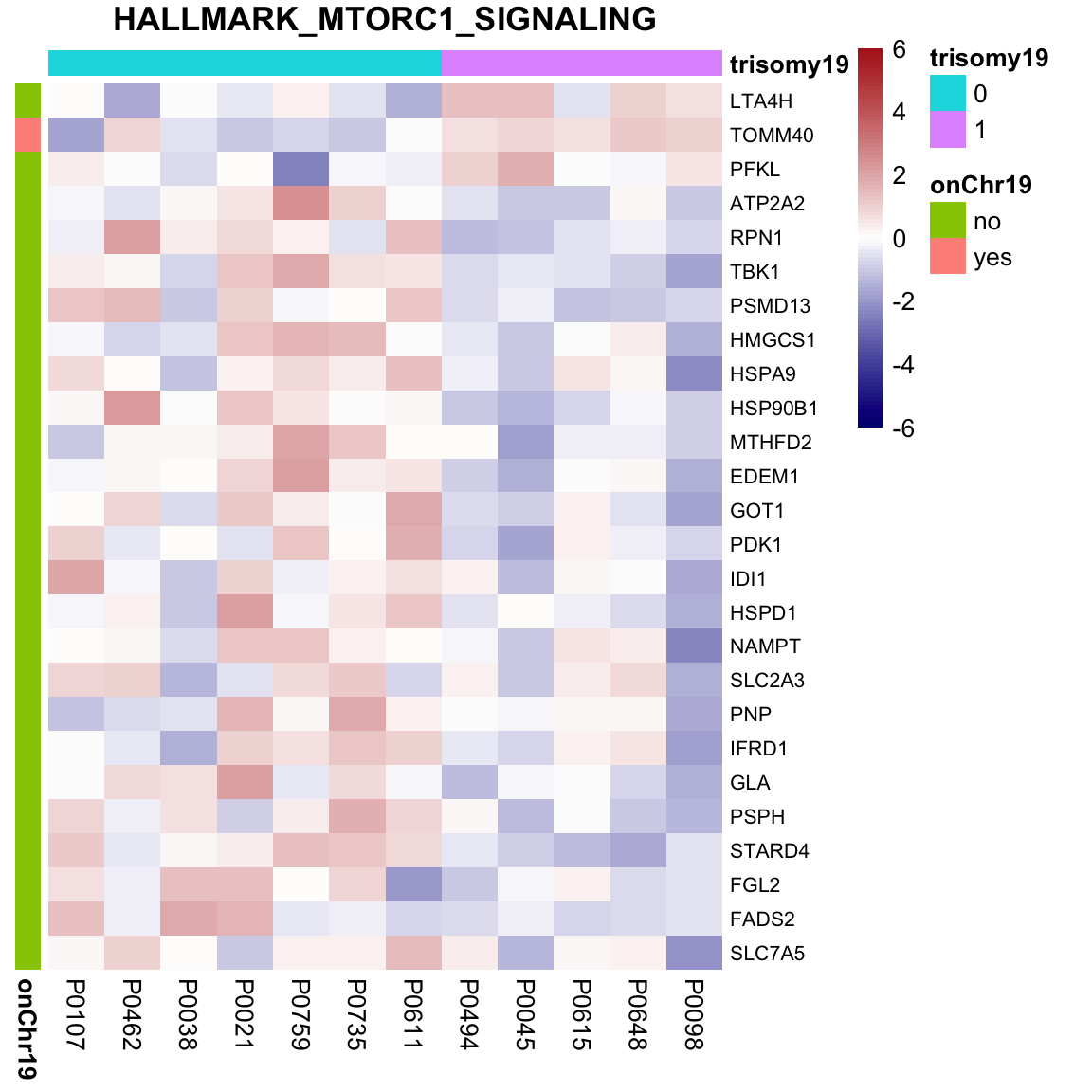
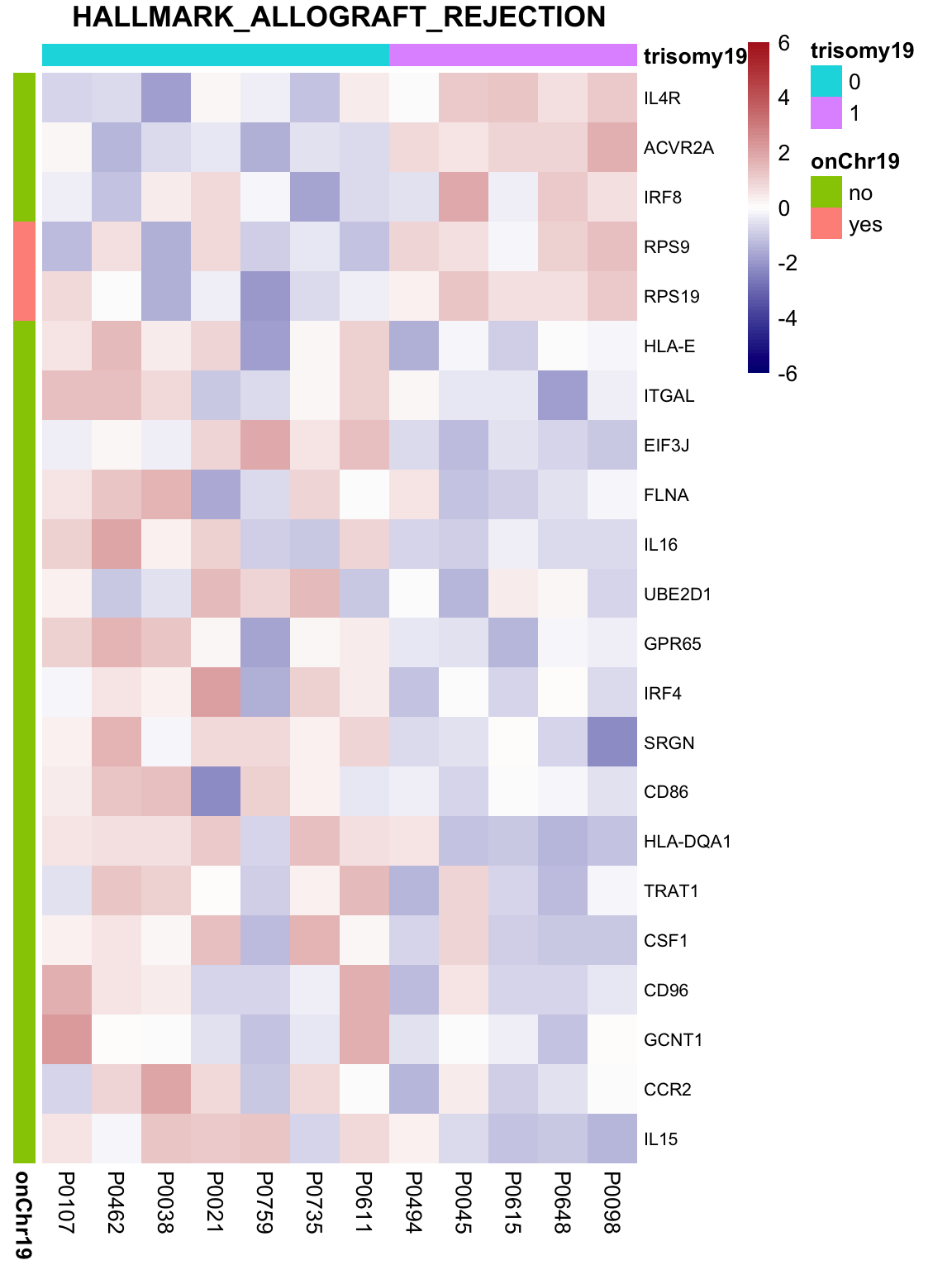
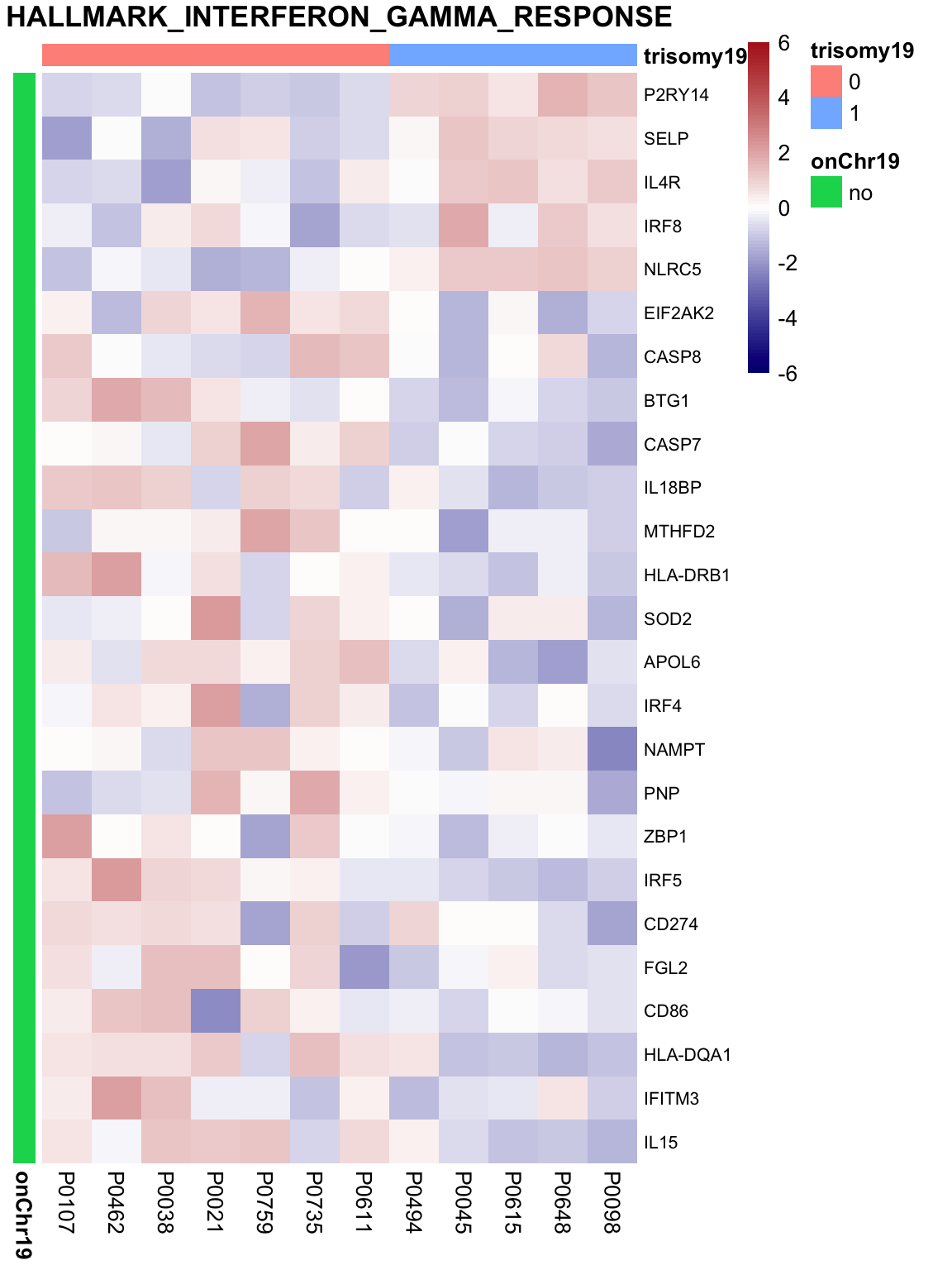
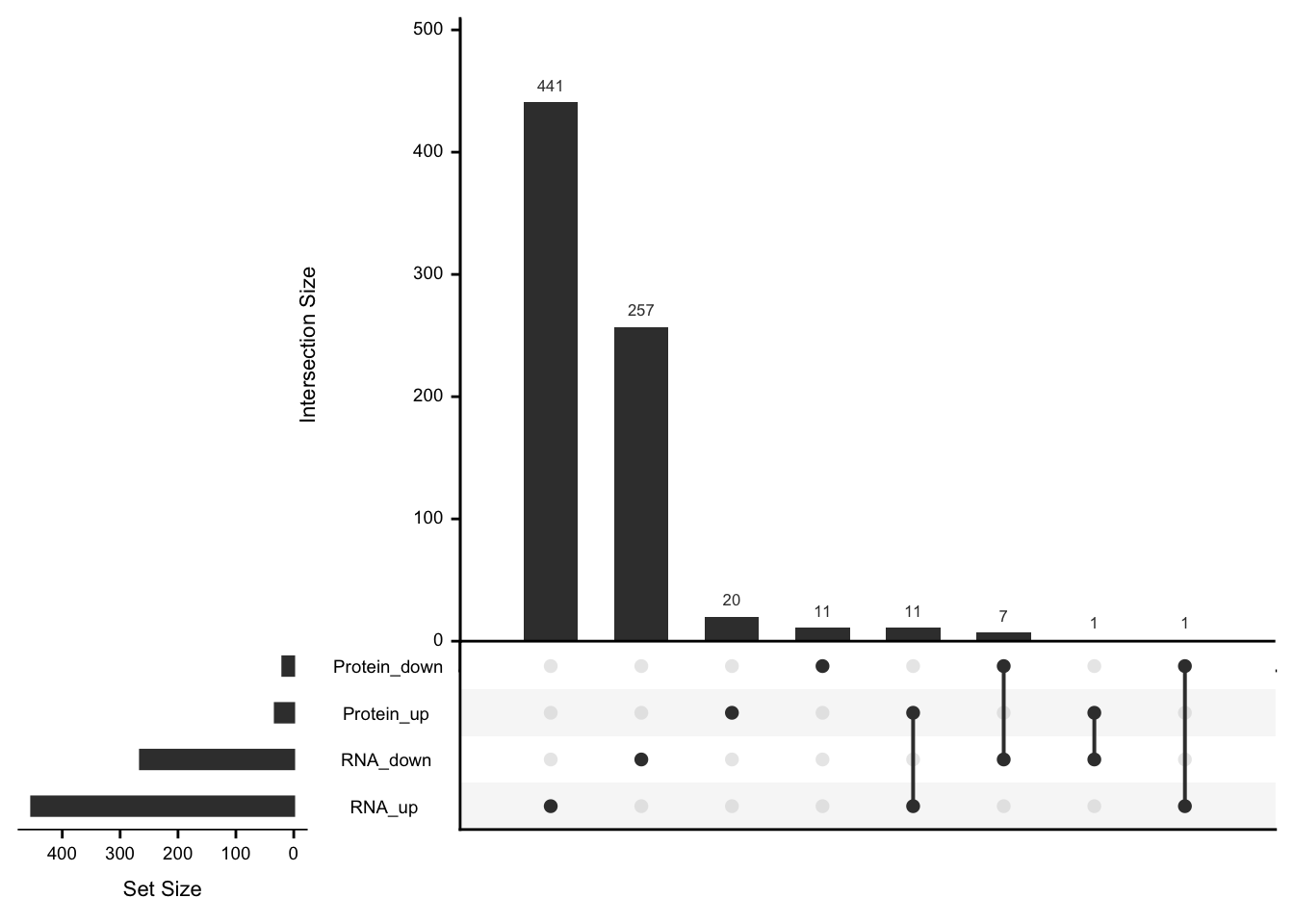
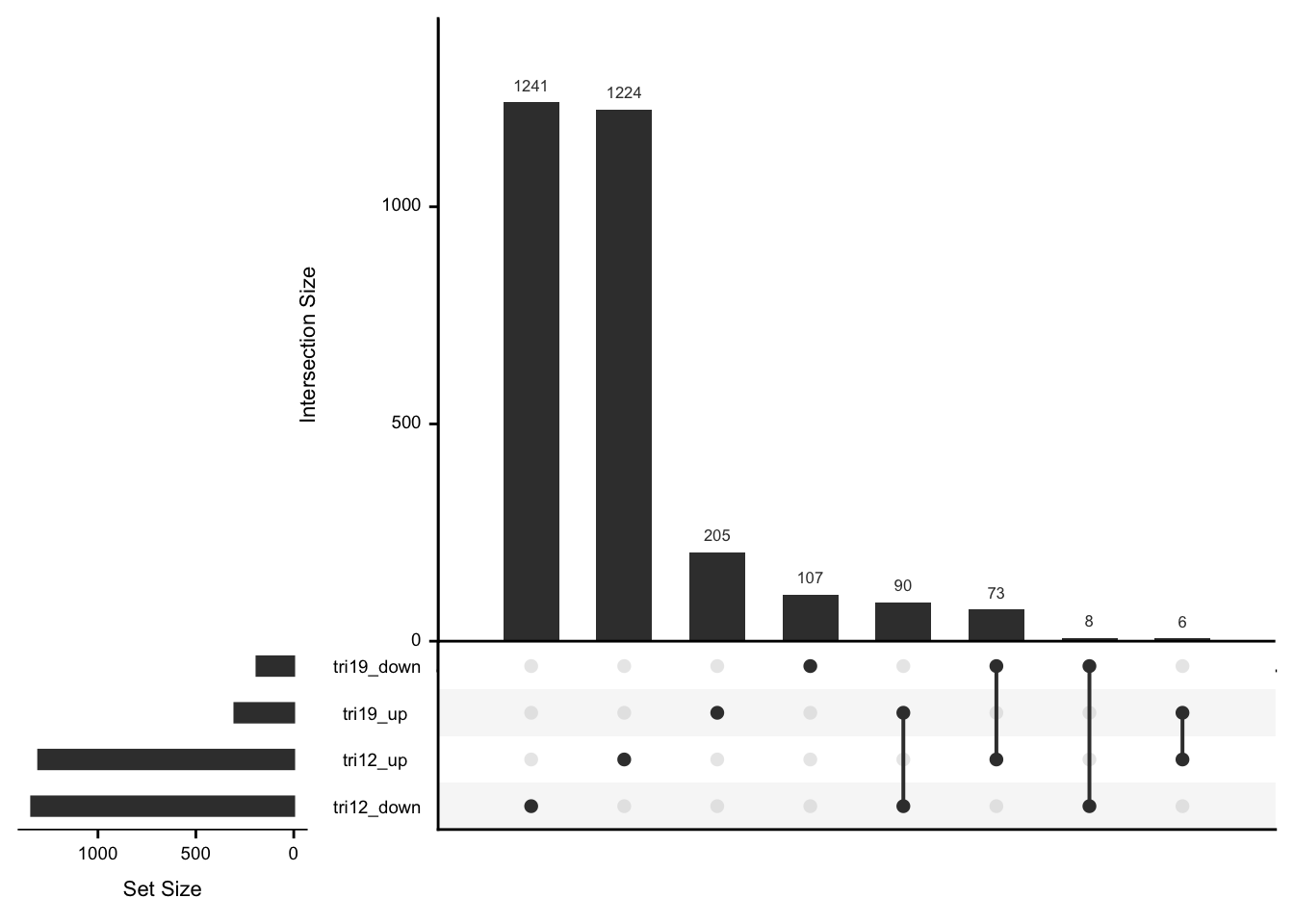
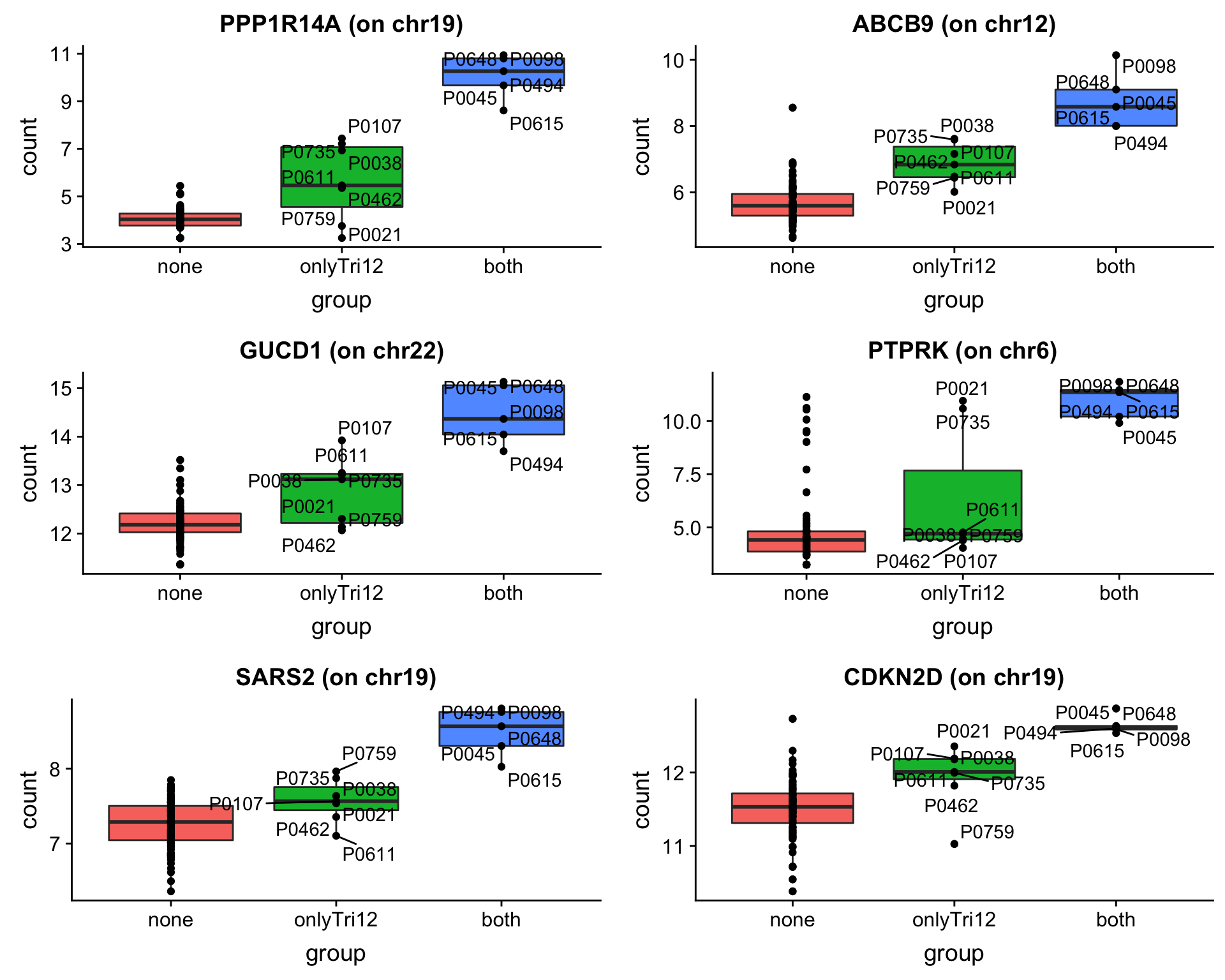
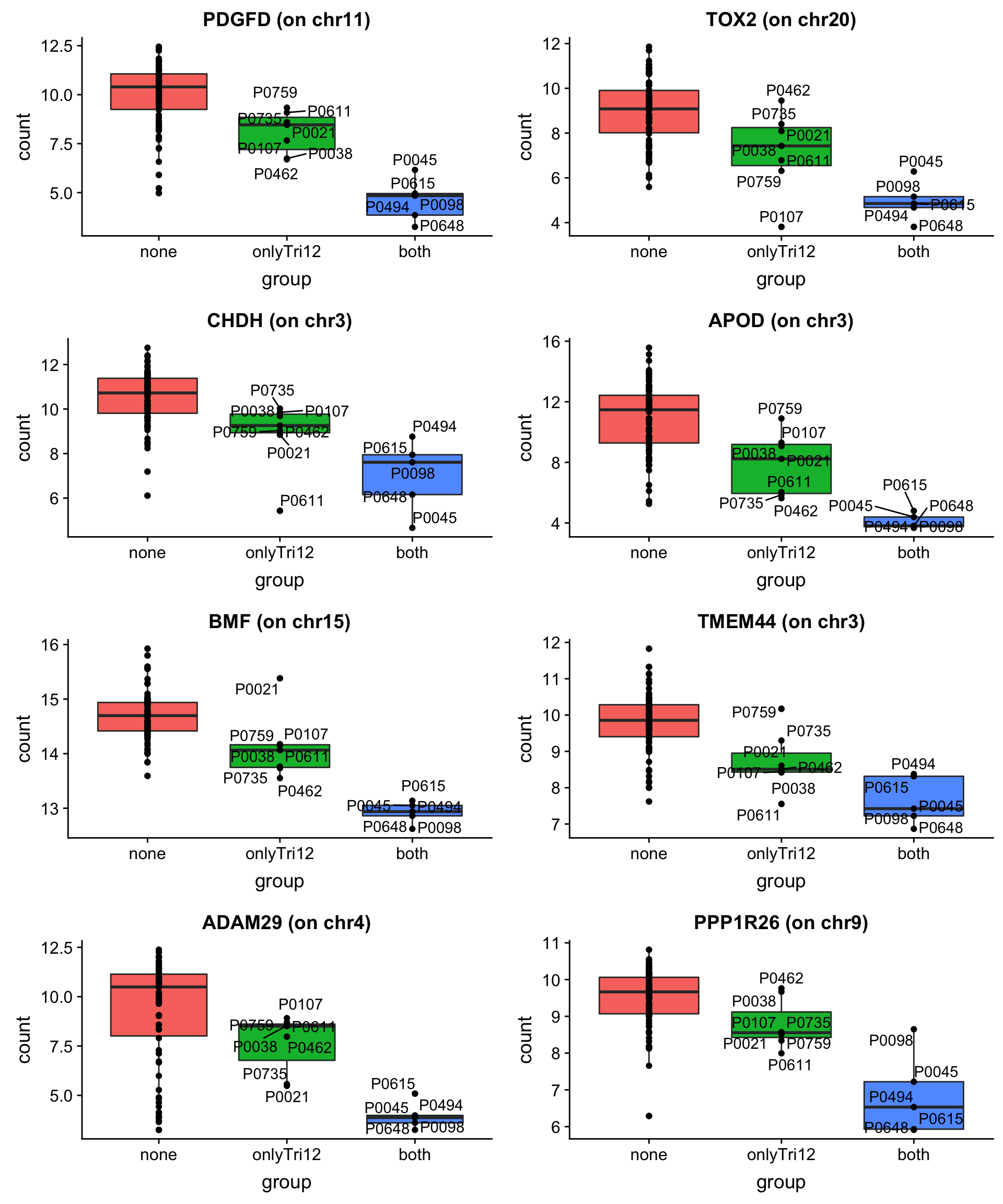
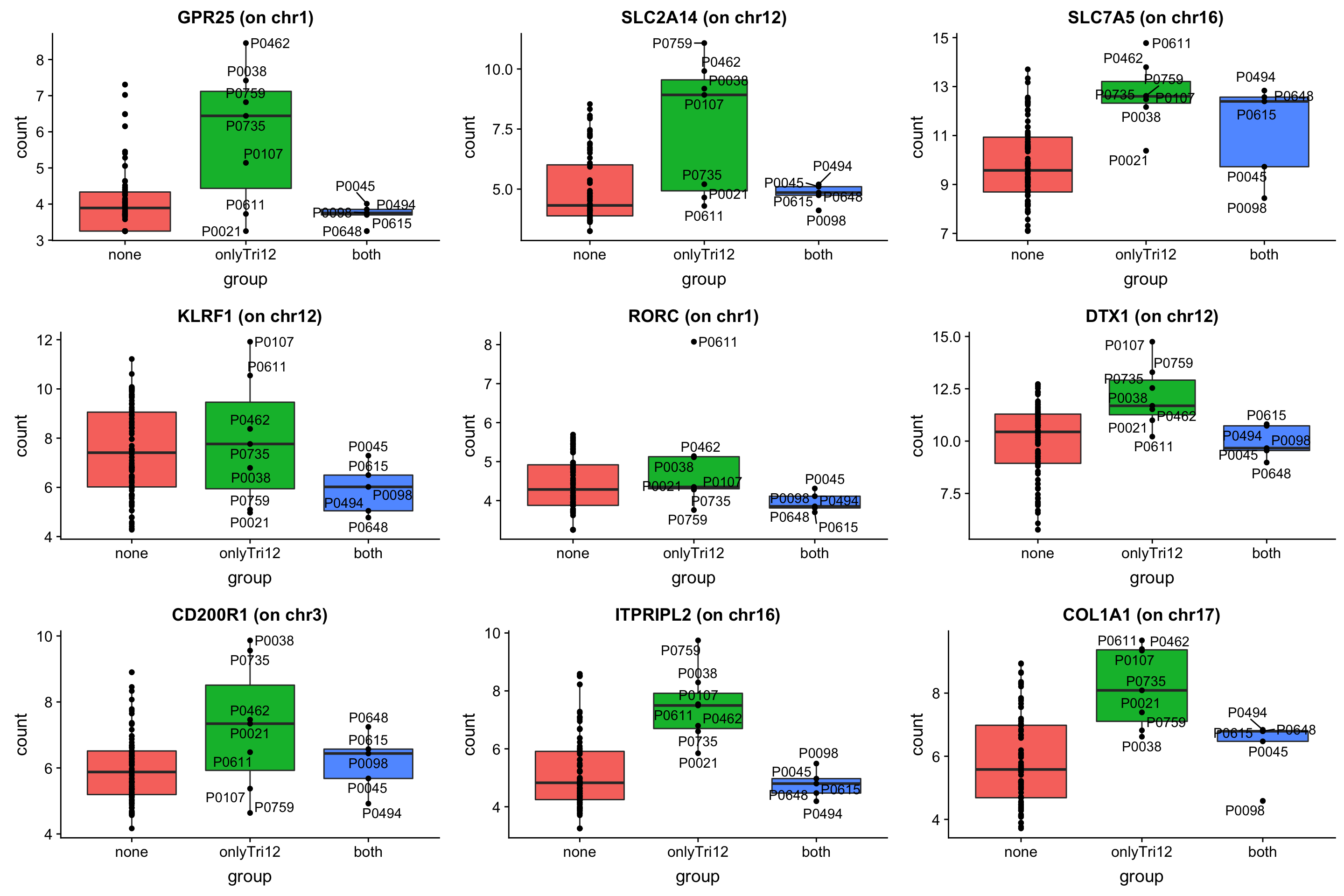
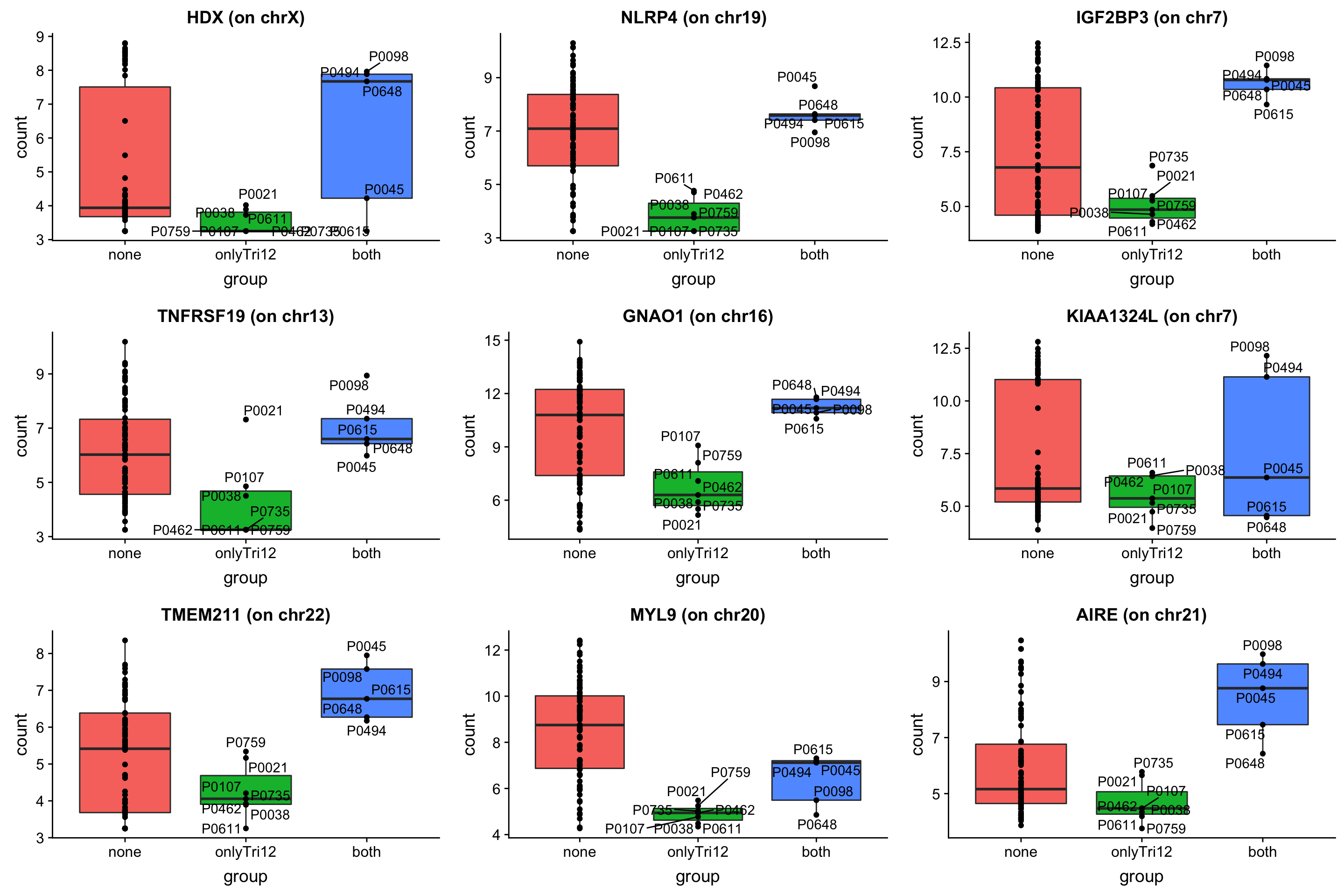
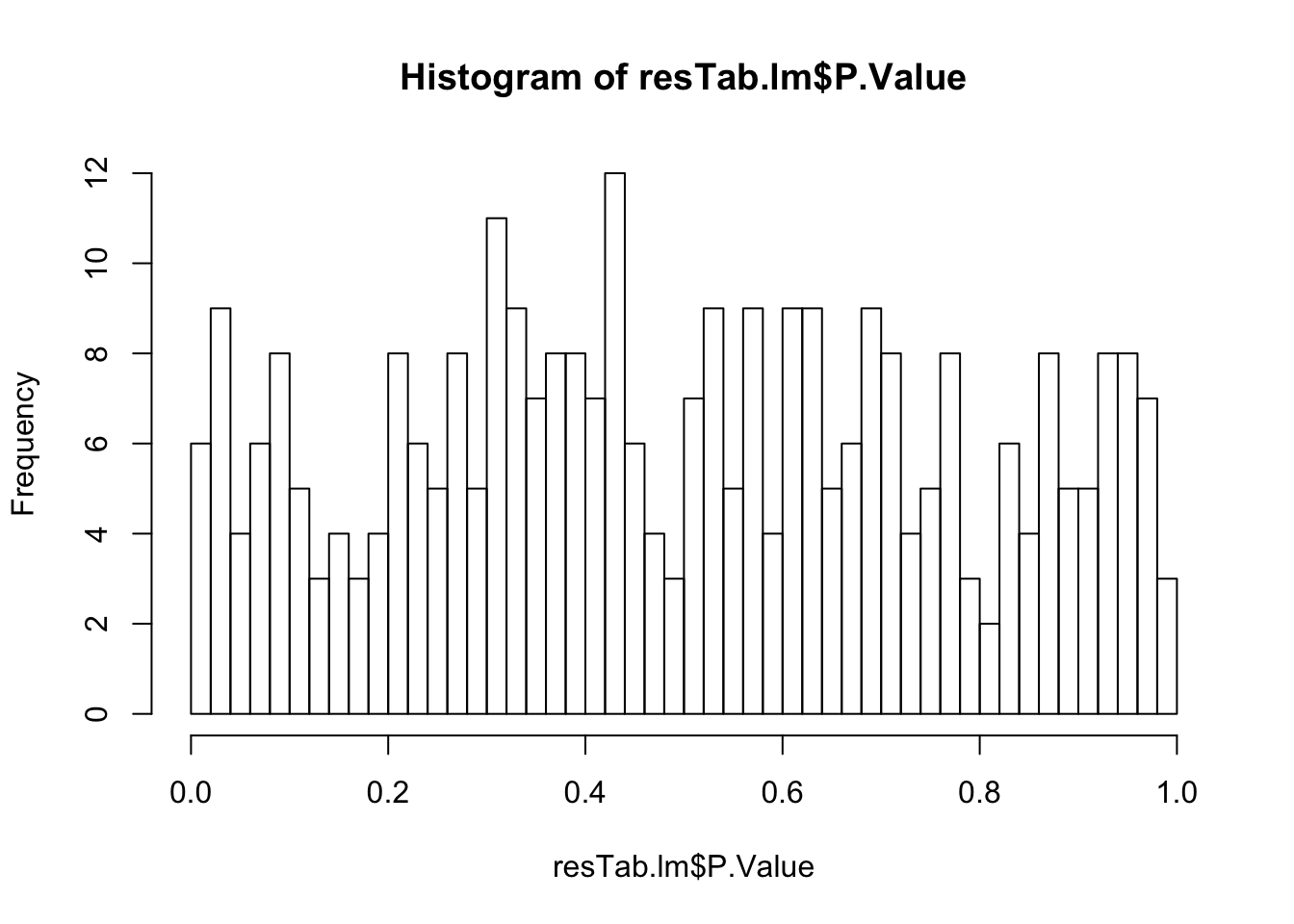 May not be many significant associations
May not be many significant associations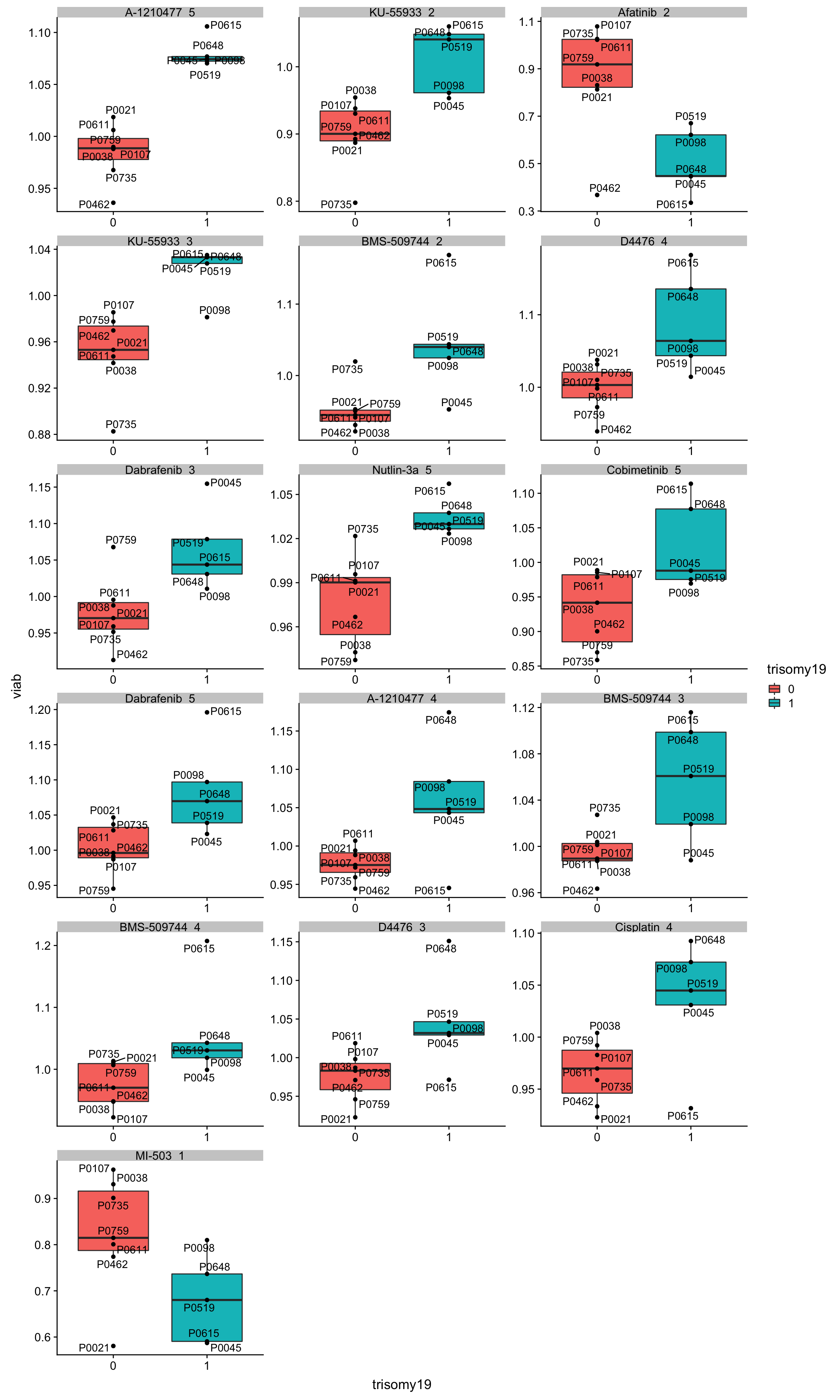 Although the individual P-Values are not very significant, but there’s a general trend that additional presence trisomy19 lead to drug resistance. But most of those associations happen at low concentrations and the overall effect size is not large. This maybe due to the baseline survival difference of samples with trisomy19.
Although the individual P-Values are not very significant, but there’s a general trend that additional presence trisomy19 lead to drug resistance. But most of those associations happen at low concentrations and the overall effect size is not large. This maybe due to the baseline survival difference of samples with trisomy19.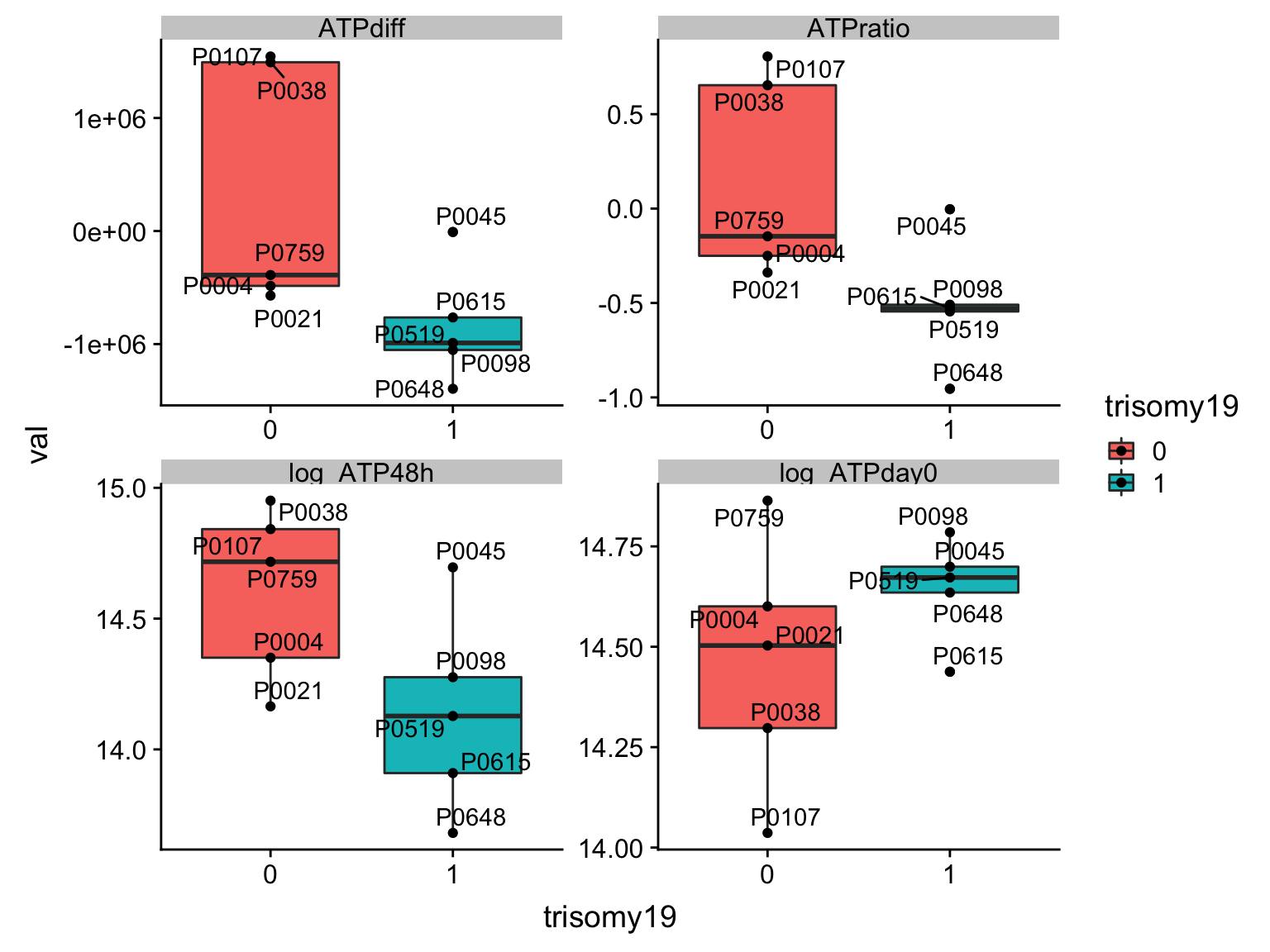 There’s a trend that samples with both trisomy19 and trisomy12 have higher day0 ATP, but tend to lose more viability during 48h culturing.
There’s a trend that samples with both trisomy19 and trisomy12 have higher day0 ATP, but tend to lose more viability during 48h culturing.