Section 8: Identifying proteomic biomarkers drug responses and clinical outcomes
Junyan Lu
2020-10-09
Last updated: 2021-03-15
Checks: 5 2
Knit directory: CLLproteomics_batch13/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200227) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- unnamed-chunk-10
- unnamed-chunk-18
- unnamed-chunk-20
- unnamed-chunk-23
To ensure reproducibility of the results, delete the cache directory manuscript_S8_drugResponse_Outcomes_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3fb50c5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/manuscript_S1_Overview_cache/
Ignored: analysis/manuscript_S3_trisomy12_cache/
Ignored: analysis/manuscript_S8_drugResponse_Outcomes_cache/
Ignored: analysis/manuscript_S9_STAT2_cache/
Ignored: code/.DS_Store
Ignored: code/.Rhistory
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/.trisomy12_norm.pdf
Untracked: analysis/STAT2splicing.Rmd
Untracked: analysis/analysisBatch2.Rmd
Untracked: analysis/bufferAnalysis.Rmd
Untracked: analysis/cohortComposition.pdf
Untracked: analysis/cohortComposition_batch2.pdf
Untracked: analysis/compareBatchClinics.Rmd
Untracked: analysis/compareBatchGenomics.Rmd
Untracked: analysis/compareTreatment.Rmd
Untracked: analysis/complexAnalysis_overall.Rmd
Untracked: analysis/corumPairs.csv
Untracked: analysis/manuscript_S1_Overview.Rmd
Untracked: analysis/manuscript_S2_genomicAssociation.Rmd
Untracked: analysis/manuscript_S3_trisomy12.Rmd
Untracked: analysis/manuscript_S4_trisomy19.Rmd
Untracked: analysis/manuscript_S5_IGHV.Rmd
Untracked: analysis/manuscript_S6_del11q.Rmd
Untracked: analysis/manuscript_S7_SF3B1.Rmd
Untracked: analysis/manuscript_S8_drugResponse_Outcomes.Rmd
Untracked: analysis/manuscript_S9_STAT2.Rmd
Untracked: analysis/patInfoTab.csv
Untracked: analysis/patInfoTab.tex
Untracked: analysis/protRNACor_eachPat.pdf
Untracked: analysis/test.pdf
Untracked: code/utils.R
Untracked: data/CNV_onChrom.RData
Untracked: data/ComplexParticipantsPubMedIdentifiers_human.txt
Untracked: data/Fig1A.png
Untracked: data/Western_blot_results_20210309_short.csv
Untracked: data/allComplexes.txt
Untracked: data/ddsrna_enc.RData
Untracked: data/exprCNV_enc.RData
Untracked: data/geneAnno.RData
Untracked: data/gmts/
Untracked: data/ic50.RData
Untracked: data/patMeta_enc.RData
Untracked: data/pepCLL_lumos_enc.RData
Untracked: data/proteins_in_complexes
Untracked: data/proteomic_explore_enc.RData
Untracked: data/proteomic_independent_enc.RData
Untracked: data/proteomic_timsTOF_enc.RData
Untracked: data/screenData_enc.RData
Untracked: data/survival_enc.RData
Untracked: output/MSH6_splicing.svg
Untracked: output/SUGP1_splicing.svg
Untracked: output/deResList.RData
Untracked: output/deResListBatch2.RData
Untracked: output/deResListRNA.RData
Untracked: output/deResList_WBC.RData
Untracked: output/deResList_batch1.RData
Untracked: output/deResList_batch3.RData
Untracked: output/deResList_timsTOF.RData
Untracked: output/dxdCLL.RData
Untracked: output/dxdCLL2.RData
Untracked: output/exprCNV.RData
Untracked: output/geneAnno.RData
Untracked: output/int_pairs.csv
Untracked: output/resOutcome_batch1.RData
Untracked: output/resOutcome_batch13.RData
Untracked: output/resOutcome_batch2.RData
Untracked: output/resOutcome_batch3.RData
Unstaged changes:
Modified: analysis/_site.yml
Deleted: analysis/analysisSF3B1.Rmd
Deleted: analysis/comparePlatforms.Rmd
Deleted: analysis/compareProteomicsRNAseq.Rmd
Deleted: analysis/correlateCLLPD.Rmd
Deleted: analysis/correlateGenomic.Rmd
Deleted: analysis/correlateGenomic_removePC.Rmd
Deleted: analysis/correlateMIR.Rmd
Deleted: analysis/correlateMethylationCluster.Rmd
Modified: analysis/index.Rmd
Deleted: analysis/predictOutcome.Rmd
Deleted: analysis/processProteomics_LUMOS.Rmd
Deleted: analysis/processProteomics_timsTOF.Rmd
Deleted: analysis/qualityControl_LUMOS.Rmd
Deleted: analysis/qualityControl_timsTOF.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Load packages and datasets
knitr::opts_chunk$set(echo = TRUE, warning = FALSE, message = FALSE, dev = c("png","pdf"))
library(limma)
library(pheatmap)
library(jyluMisc)
library(survival)
library(survminer)
library(maxstat)
library(glmnet)
library(SummarizedExperiment)
library(cowplot)
library(tidyverse)
load("../data/patMeta_enc.RData")
load("../data/ddsrna_enc.RData")
load("../data/proteomic_explore_enc.RData")
load("../output/deResList.RData") #precalculated differential expression
load("../data/survival_enc.RData")
load("../data/screenData_enc.RData")
#protCLL <- protCLL[rowData(protCLL)$uniqueMap,]
source("../code/utils.R")Correlations between protein abundance and drug response
Preprocess datasets
Drug screening data from 1000CPS: low quality samples after QC are removed; the edge effect corrected viability values are used
viabMat <- screenData %>% filter(!lowQuality, ! Drug %in% c("DMSO","PBS"), patientID %in% colnames(protCLL)) %>%
group_by(patientID, Drug) %>% summarise(viab = mean(normVal.adj.cor_auc)) %>%
spread(key = patientID, value = viab) %>%
data.frame(stringsAsFactors = FALSE) %>% column_to_rownames("Drug") %>%
as.matrix()Proteomics data
proMat <- assays(protCLL)[["count"]]
proMat <- proMat[,colnames(viabMat)]
#Remove proteins without much variance (to lower multi-testing burden)
#sds <- genefilter::rowSds(proMat,na.rm=TRUE)
#proMat <- proMat[sds > genefilter::shorth(sds),]How many samples have both proteomics data and CPS1000 screen data
ncol(proMat)[1] 82Association test
resTab.auc <- lapply(rownames(viabMat),function(drugName) {
viab <- viabMat[drugName, ]
batch <- protCLL[,colnames(viabMat)]$batch
designMat <- model.matrix(~1+viab+batch)
fit <- lmFit(proMat, designMat)
fit2 <- eBayes(fit)
corRes <- topTable(fit2, number ="all", adjust.method = "BH", coef = "viab") %>% rownames_to_column("id") %>%
mutate(symbol = rowData(protCLL[id,])$hgnc_symbol, Drug = drugName)
}) %>% bind_rows() %>% mutate(adj.P.Val = p.adjust(P.Value, method = "BH")) %>% arrange(P.Value)Associations test with blocking for WBC counts
resTab.block <- lapply(rownames(viabMat),function(drugName) {
viab <- viabMat[drugName, ]
batch <- protCLL[,colnames(viabMat)]$batch
leukCount <- log10(sampleTab[match(colnames(viabMat), sampleTab$encID),]$leukCount)
designMat <- model.matrix(~1+viab+batch+leukCount)
fit <- lmFit(proMat, designMat)
fit2 <- eBayes(fit)
corRes <- topTable(fit2, number ="all", adjust.method = "BH", coef = "viab") %>% rownames_to_column("id") %>%
mutate(symbol = rowData(protCLL[id,])$hgnc_symbol, Drug = drugName)
}) %>% bind_rows() %>% mutate(adj.P.Val = p.adjust(P.Value, method = "BH")) %>% arrange(P.Value)Compare results with and without blocking for WBC
tabNoBlock <- resTab.auc %>% mutate(ifSig = adj.P.Val <= 0.05) %>%
select(id, Drug, P.Value, ifSig)
tabBlock <- resTab.block %>% mutate(ifSig.block = adj.P.Val <= 0.05, P.Value.block = P.Value) %>%
select(id, Drug, P.Value.block, ifSig.block)
compareTab <- left_join(tabNoBlock, tabBlock, by = c("id","Drug")) %>%
mutate(group = case_when(
ifSig & ifSig.block ~ "significant in both",
ifSig & !ifSig.block ~ "significant only without blocking",
!ifSig & ifSig.block ~ "significant only with blocking",
TRUE ~ "not significant in both"
))
ggplot(compareTab, aes(x=-log10(P.Value), y=-log10(P.Value.block), col = group)) +
geom_point(alpha=0.5) +
xlab("-log10(P value) without blocking") + ylab("-log10 (P value) with blocking for WBC counts") +
scale_color_manual(values = c("grey80", colList[2], colList[1], colList[3]), name = "") +
geom_abline(slope = 1, linetype ="dashed") +
ggtitle("Associations with drug responses") +
theme_full +
theme(legend.position = c(0.7,0.2))
Number of associations in each catagory
tt <- table(compareTab$group)
tt
not significant in both significant in both
208175 524
significant only with blocking significant only without blocking
38 45 A percentage of consistent results
(tt[1] + tt[2])/sum(tt)*100not significant in both
99.96025 Bar plot to show the number of associations (10% FDR)
#Select significant associations (10% FDR)
resTab.sig <- filter(resTab.auc, adj.P.Val <= 0.05) %>%
select(Drug, symbol, id,logFC, P.Value, adj.P.Val)
plotTab <- resTab.sig %>% group_by(Drug) %>%
summarise(n = length(id)) %>% ungroup()
ordTab <- group_by(plotTab, Drug) %>% summarise(total = sum(n)) %>%
arrange(desc(total))
plotTab <- mutate(plotTab, Drug = factor(Drug, levels = ordTab$Drug)) %>%
filter(n>0)
drugBar <- ggplot(plotTab, aes(x=Drug, y = n)) + geom_bar(stat="identity",fill=colList[4]) +
geom_text(aes(label = paste0("n=", n)),vjust=-1,col=colList[1]) +
ylim(0,200)+ annotate("text", label = "Number of associations (10% FDR)", x=Inf, y=Inf,hjust=1, vjust=1, size=6)+
theme_half + theme(axis.text.x = element_text(angle = 90, hjust=1, vjust=0.5)) +
ylab("Number") + xlab("")
drugBar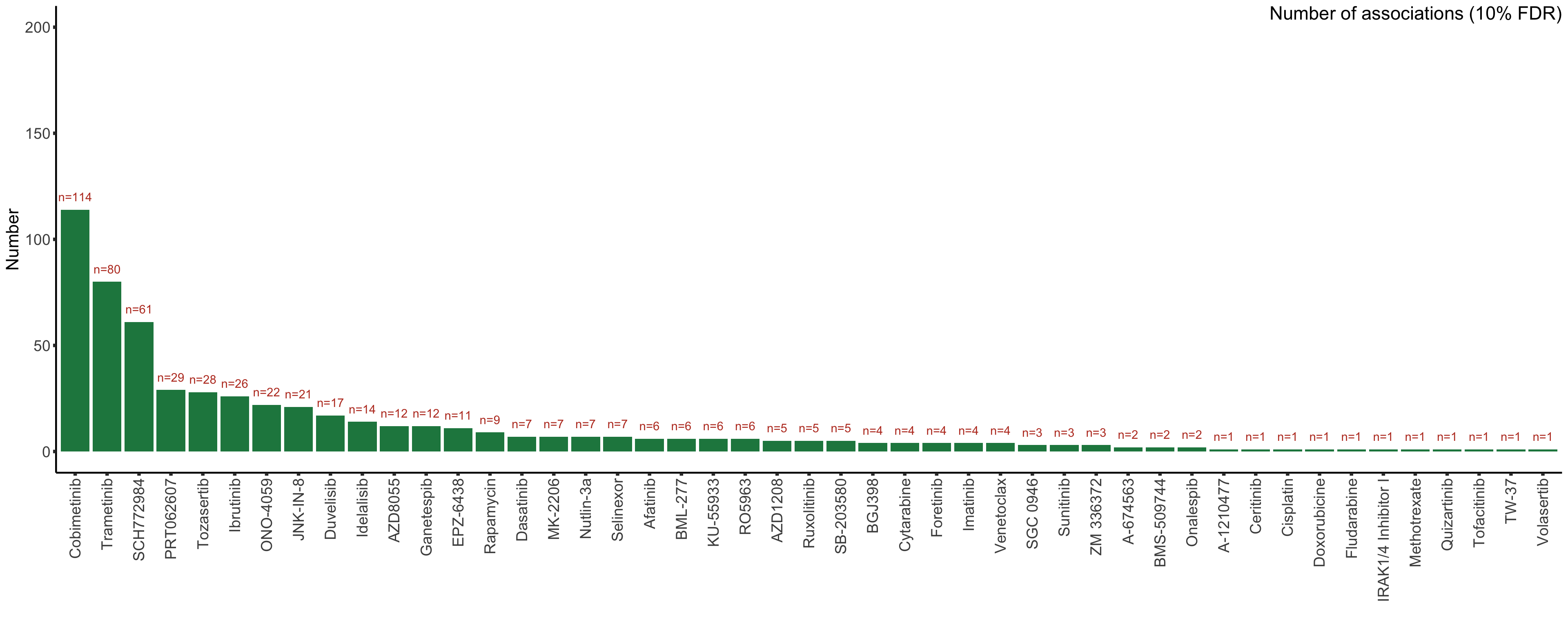
Table of significant associations (10% FDR)
resTab.sig %>% mutate_if(is.numeric, formatC, digits=2, format= "e") %>%
DT::datatable()Correlation plot of selected candidates
plotDrugCorScatter <- function(inputTab, x, y, x_lab = "X", y_lab = "Y", title = "",
col = NULL, showR2 = TRUE, annoPos = "right",
dotCol = colList, textCol="darkred") {
#prepare table for plotting
plotTab <- tibble(x = inputTab[[x]],y=inputTab[[y]])
if (!is.null(col)) plotTab <- mutate(plotTab, status = inputTab[[col]])
plotTab <- filter(plotTab, !is.na(x), !is.na(y))
#prepare annotation values
corRes <- cor.test(plotTab$x, plotTab$y)
pval <- formatNum(corRes$p.value, digits = 1, format = "e")
Rval <- formatNum(corRes$estimate, digits = 1, format = "e")
R2val <- formatNum(corRes$estimate^2, digits = 1, format = "e")
Nval <- nrow(plotTab)
annoP <- bquote(italic("P")~"="~.(pval))
if (showR2) {
annoCoef <- bquote(R^2~"="~.(R2val))
} else {
annoCoef <- bquote(R~"="~.(Rval))
}
annoN <- bquote(N~"="~.(Nval))
corPlot <- ggplot(plotTab, aes(x = x, y = y))
if (!is.null(col)) {
corPlot <- corPlot + geom_point(aes(fill = status), shape =21, size =3) +
scale_fill_manual(values = dotCol)
} else {
corPlot <- corPlot + geom_point(fill = dotCol[1], shape =21, size=3)
}
corPlot <- corPlot + geom_smooth(formula = y~x,method = "lm", se=FALSE, color = "grey50", linetype ="dashed" )
if (annoPos == "right") {
corPlot <- corPlot + annotate("text", x = max(plotTab$x), y = Inf, label = annoN,
hjust=1, vjust =2, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = max(plotTab$x), y = Inf, label = annoP,
hjust=1, vjust =4, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = max(plotTab$x), y = Inf, label = annoCoef,
hjust=1, vjust =6, size = 5, parse = FALSE, col= textCol)
} else if (annoPos== "left") {
corPlot <- corPlot + annotate("text", x = min(plotTab$x), y = Inf, label = annoN,
hjust=0, vjust =2, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = min(plotTab$x), y = Inf, label = annoP,
hjust=0, vjust =4, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = min(plotTab$x), y = Inf, label = annoCoef,
hjust=0, vjust =6, size = 5, parse = FALSE, col= textCol)
}
corPlot <- corPlot + ylab(y_lab) + xlab(x_lab) + ggtitle(title) +
scale_y_continuous(labels = scales::number_format(accuracy = 0.1)) +
scale_x_continuous(labels = scales::number_format(accuracy = 0.1)) +
theme_full
corPlot
}proMat.combat <- assays(protCLL)[["count_combat"]]
proMat.combat <- proMat.combat[,colnames(viabMat)]
pairList <- list(c("Cobimetinib","STAT2"),c("Venetoclax","PGD"),c("Idelalisib", "HEBP2"))
plotList <- lapply(pairList, function(pair) {
textCol <- "darkred"
drugName <- pair[1]
proteinName <- pair[2]
id <- rownames(protCLL)[match(proteinName, rowData(protCLL)$hgnc_symbol)]
plotTab <- tibble(patID = colnames(viabMat),
viab = viabMat[drugName,],
expr = proMat.combat[id,]) %>%
mutate(IGHV = protCLL[,patID]$IGHV.status,
trisomy12 = protCLL[,patID]$trisomy12) %>%
mutate(trisomy12 = ifelse(trisomy12 ==1,"yes","no")) %>%
filter(!is.na(viab),!is.na(expr))
pval <- formatNum(filter(resTab.sig, Drug == drugName, symbol == proteinName)$P.Value, digits = 1, format = "e")
Rval <- sprintf("%1.2f",cor(plotTab$viab, plotTab$expr))
Nval <- nrow(plotTab)
annoP <- bquote(italic("P")~"="~.(pval))
annoN <- bquote(N~"="~.(Nval))
annoCoef <- bquote(R~"="~.(Rval))
corPlot <- ggplot(plotTab, aes(x = viab, y = expr)) +
geom_point(aes(col = trisomy12, shape = IGHV), size=5) +
scale_shape_manual(values = c(M = 19, U = 1)) +
scale_color_manual(values = c(yes = colList[2], no = colList[3])) +
geom_smooth(formula = y~x,method = "lm", se=FALSE, color = "grey50", linetype ="dashed" ) +
ggtitle(sprintf("%s ~ %s", drugName, proteinName)) +
ylab("Protein expression") + xlab("Viability after treatment") +
theme_full +
theme(legend.position = "bottom")
if (Rval < 0) annoPos <- "right" else annoPos <- "left"
if (annoPos == "right") {
corPlot <- corPlot + annotate("text", x = max(plotTab$viab), y = Inf, label = annoN,
hjust=1, vjust =2, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = max(plotTab$viab), y = Inf, label = annoP,
hjust=1, vjust =4, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = max(plotTab$viab), y = Inf, label = annoCoef,
hjust=1, vjust =6, size = 5, parse = FALSE, col= textCol)
} else if (annoPos== "left") {
corPlot <- corPlot + annotate("text", x = min(plotTab$viab), y = Inf, label = annoN,
hjust=0, vjust =2, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = min(plotTab$viab), y = Inf, label = annoP,
hjust=0, vjust =4, size = 5, parse = FALSE, col= textCol) +
annotate("text", x = min(plotTab$viab), y = Inf, label = annoCoef,
hjust=0, vjust =6, size = 5, parse = FALSE, col= textCol)
}
corPlot <- corPlot + ylab("Protein expression") + xlab("Viability after treatment") +
scale_y_continuous(labels = scales::number_format(accuracy = 0.1)) +
scale_x_continuous(labels = scales::number_format(accuracy = 0.1))
corPlot
})
drugCor <- cowplot::plot_grid(plotlist = plotList, ncol =3)
drugCor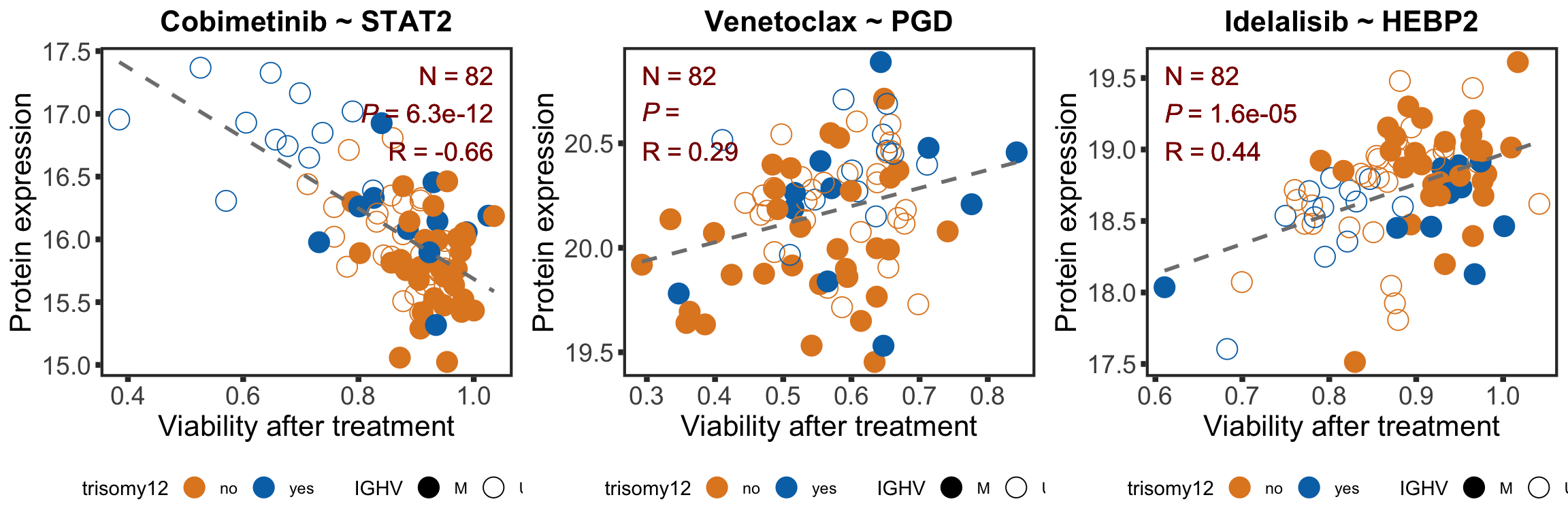
Association test with blocking for IGHV and trisomy12
testList <- filter(resTab.auc, adj.P.Val <= 0.05)
resTab.auc.block <- lapply(seq(nrow(testList)),function(i) {
pair <- testList[i,]
expr <- proMat[pair$id,]
viab <- viabMat[pair$Drug, ]
ighv <- protCLL[,colnames(viabMat)]$IGHV.status
tri12 <- protCLL[,colnames(viabMat)]$trisomy12
batch <- protCLL[,colnames(viabMat)]$batch
res <- anova(lm(viab~ighv+tri12+batch+expr))
data.frame(id = pair$id, P.Value = res["expr",]$`Pr(>F)`, symbol = pair$symbol,
Drug = pair$Drug,
P.Value.IGHV = res["ighv",]$`Pr(>F)`,P.Value.trisomy12 = res["tri12",]$`Pr(>F)`,
P.Value.noBlock = pair$P.Value,
stringsAsFactors = FALSE)
}) %>% bind_rows() %>% mutate(adj.P.Val = p.adjust(P.Value, method = "BH")) %>% arrange(P.Value)Assocations that are still significant
resTab.sig %>% mutate_if(is.numeric, formatC, digits=2, format= "e") %>%
DT::datatable()The above mentioned pairs are still significant.
Variance explained
Test whether the protein expression can explain additional variance in drug response compared to genetic alone
Prepare genomics
geneMat <- patMeta[match(colnames(proMat), patMeta$Patient.ID),] %>%
select(Patient.ID, IGHV.status, del11q:U1) %>%
mutate_if(is.factor, as.character) %>% mutate(IGHV.status = ifelse(IGHV.status == "M", 1,0)) %>%
mutate_at(vars(-Patient.ID), as.numeric) %>% #assign a few unknown mutated cases to wildtype
mutate_all(replace_na,0) %>%
data.frame() %>% column_to_rownames("Patient.ID")
geneMat <- geneMat[,apply(geneMat,2, function(x) sum(x %in% 1, na.rm = TRUE))>=5] %>% as.matrix()Genes that will be included in the multivariate model
colnames(geneMat) [1] "IGHV.status" "del11q" "del13q" "del17p" "trisomy12"
[6] "trisomy19" "ATM" "BRAF" "DDX3X" "EGR2"
[11] "NOTCH1" "SF3B1" "TP53" compareR2 <- function(Drug, protName, geneMat) {
viab <- viabMat[Drug,]
protID <- unique(filter(resTab.sig, symbol == protName)$id)
expr <- proMat[protID,]
tabGene <- data.frame(geneMat)
tabGene[["viab"]] <- viab
tabCom <- tabGene
tabCom[[protName]] <- expr
r2Prot <- summary(lm(viab~expr))$r.squared
r2Gene <- summary(lm(viab~., data=tabGene))$r.squared
r2Com <- summary(lm(viab~., data=tabCom))$r.squared
plotTab <- tibble(model = c("genetics",paste0(protName, " expression"),sprintf("genetics + \n%s expression",protName)),
R2 = c(r2Gene, r2Prot, r2Com))
ggplot(plotTab, aes(x=model, y = R2)) + geom_bar(aes(fill = model),stat="identity", width=0.6) + coord_flip() +
ggtitle(Drug) + ylab("Variance expalained (R2)") + xlab("") +
scale_fill_manual(values = colList[4:6]) + theme_full +theme(legend.position = "none")
}plotList <- lapply(pairList, function(p) {
compareR2(p[1],p[2],geneMat)
})
plot_grid(plotlist= plotList, ncol= 1)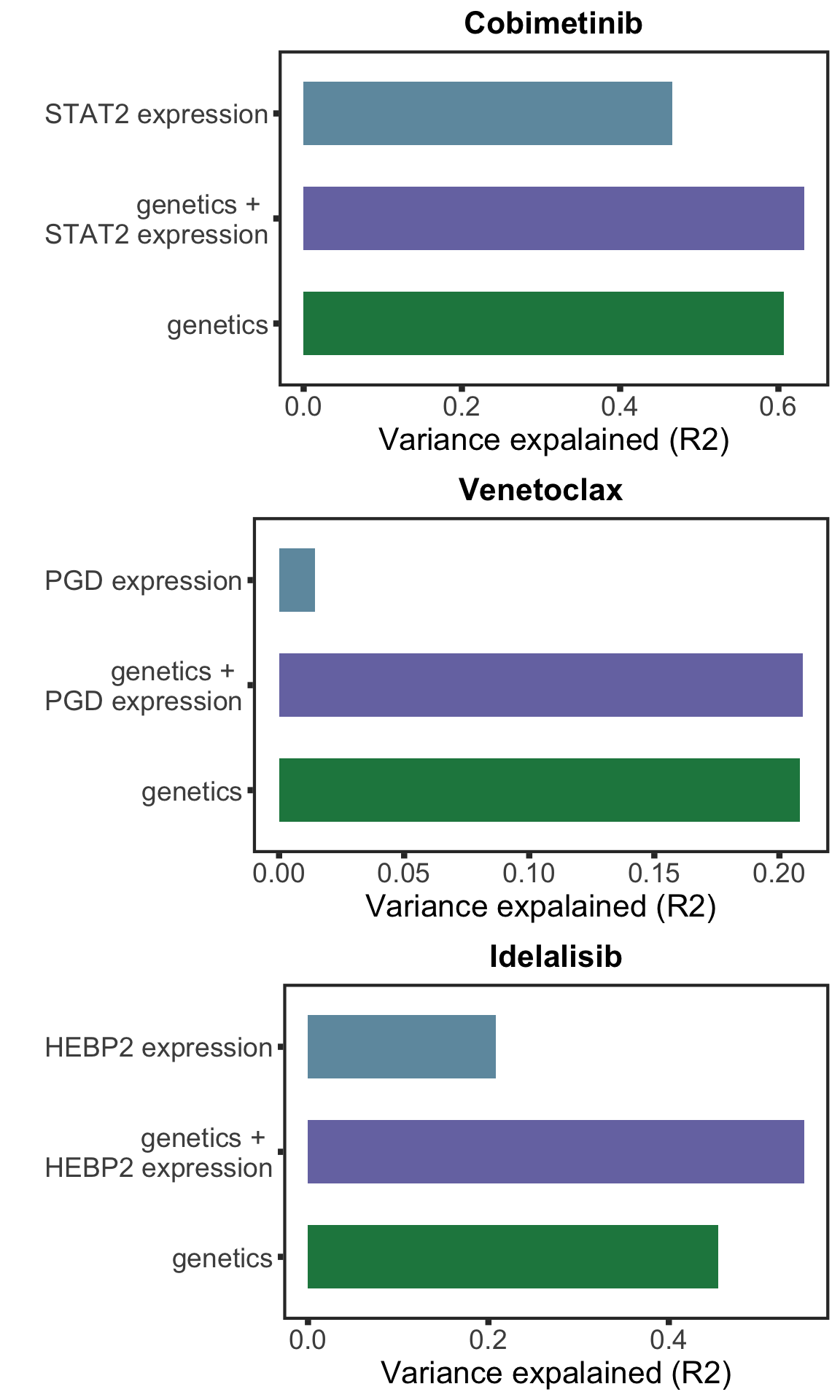 Based on the plot of R2 values, including protein expression in multi-variate model could explain additional variance in drug responses compared to genetic alone. For Venetoclax, using a single protein expression value of PGD already explain more variance than genetics.
Based on the plot of R2 values, including protein expression in multi-variate model could explain additional variance in drug responses compared to genetic alone. For Venetoclax, using a single protein expression value of PGD already explain more variance than genetics.
stat2Bar <- plotList[[1]]
stat2Bar
Protein markers for clincial outcomes
Uni-variate model to identify proteins associated with outcomes
protCLL.sub <- protCLL[!rowData(protCLL)$chromosome_name %in% c("X","Y"),]
protMat <- assays(protCLL.sub)[["count_combat"]]
survTab <- survT %>%
select(patID, OS, died, TTT, treatedAfter, age,sex) %>%
dplyr::rename(patientID = patID) %>%
filter(patientID %in% colnames(protMat))TTT events
table(survTab$treatedAfter)
FALSE TRUE
44 46 OS events
table(survTab$died)
FALSE TRUE
80 11 uniRes.ttt <- lapply(rownames(protMat), function(n) {
testTab <- mutate(survTab, expr = protMat[n, patientID])
com(testTab$expr, testTab$TTT, testTab$treatedAfter, TRUE) %>%
mutate(id = n)
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH")) %>%
arrange(p) %>% mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
mutate(outcome = "TTT")
uniRes.os <- lapply(rownames(protMat), function(n) {
testTab <- mutate(survTab, expr = protMat[n, patientID])
com(testTab$expr, testTab$OS, testTab$died, TRUE) %>%
mutate(id = n)
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH")) %>%
arrange(p) %>% mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
mutate(outcome = "OS")
uniRes <- bind_rows(uniRes.ttt, uniRes.os) %>%
mutate(p.adj = p.adjust(p, method = "BH"))A table showing significant associations
uniRes %>% filter(p.adj <= 0.05) %>% mutate_if(is.numeric, formatC, digits=2,format="e") %>%
select(name, p, HR, p.adj, outcome) %>% DT::datatable()Blocking for WBC count using multi-variate model
Multi-variate test
uniRes.ttt.block <- lapply(rownames(protMat), function(n) {
riskTab <- select(survTab, patientID) %>%
mutate(leukCount = log10(sampleTab[match(patientID, sampleTab$encID),]$leukCount)) %>%
mutate(protExpr = protMat[n,patientID])
fullModel <- runCox(survTab, riskTab, "TTT","treatedAfter")
res <- summary(fullModel)
tibble(id = n, p = res$coefficients["protExpr",5],)
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH"))
uniRes.os.block <- lapply(rownames(protMat), function(n) {
riskTab <- select(survTab, patientID) %>%
mutate(leukCount = log10(sampleTab[match(patientID, sampleTab$encID),]$leukCount)) %>%
mutate(protExpr = protMat[n,patientID])
fullModel <- runCox(survTab, riskTab, "OS","died")
res <- summary(fullModel)
tibble(id = n, p = res$coefficients["protExpr",5],)
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH"))Compare results from blocking and non-blocking
TTT
tabNoBlock <- uniRes.ttt %>% mutate(ifSig = p.adj <= 0.05) %>%
select(id, p, ifSig)
tabBlock <- uniRes.ttt.block %>% mutate(ifSig.block = p.adj <= 0.05, p.block = p) %>%
select(id, p.block, ifSig.block)
compareTab <- left_join(tabNoBlock, tabBlock, by = "id") %>%
mutate(group = case_when(
ifSig & ifSig.block ~ "significant in both",
ifSig & !ifSig.block ~ "significant only without blocking",
!ifSig & ifSig.block ~ "significant only with blocking",
TRUE ~ "not significant in both"
))
ggplot(compareTab, aes(x=-log10(p), y=-log10(p.block), col = group)) +
geom_point(alpha=0.5) +
xlab("-log10(P value) without blocking") + ylab("-log10 (P value) with blocking for WBC counts") +
scale_color_manual(values = c("grey80", colList[2], colList[1], colList[3]), name = "") +
geom_abline(slope = 1, linetype ="dashed") +
ggtitle("Associations with TTT") +
theme_full +
theme(legend.position = c(0.7,0.2))
Number of associations in each catagory
tt <- table(compareTab$group)
tt
not significant in both significant in both
2859 189
significant only with blocking significant only without blocking
104 53 A percentage of consistent results
(tt[1] + tt[2])/sum(tt)*100not significant in both
95.1014 OS
tabNoBlock <- uniRes.os %>% mutate(ifSig = p.adj <= 0.05) %>%
select(id, p, ifSig)
tabBlock <- uniRes.os.block %>% mutate(ifSig.block = p.adj <= 0.05, p.block = p) %>%
select(id, p.block, ifSig.block)
compareTab <- left_join(tabNoBlock, tabBlock, by = "id") %>%
mutate(group = case_when(
ifSig & ifSig.block ~ "significant in both",
ifSig & !ifSig.block ~ "significant only without blocking",
!ifSig & ifSig.block ~ "significant only with blocking",
TRUE ~ "not significant in both"
))
ggplot(compareTab, aes(x=-log10(p), y=-log10(p.block), col = group)) +
geom_point(alpha=0.5) +
xlab("-log10(P value) without blocking") + ylab("-log10 (P value) with blocking for WBC counts") +
scale_color_manual(values = c("grey80", colList[2], colList[1], colList[3]), name = "") +
geom_abline(slope = 1, linetype ="dashed") +
ggtitle("Associations with OS") +
theme_full +
theme(legend.position = c(0.7,0.2))
Number of associations in each catagory
tt <- table(compareTab$group)
tt
not significant in both significant only without blocking
3204 1 A percentage of consistent results
(tt[1] + tt[2])/sum(tt)*100not significant in both
100 Selection protein markers independent of known risks using multi-vairate model
Prepare data
#table of known risks
riskTab <- select(survTab, patientID, age, sex) %>%
left_join(patMeta[,c("Patient.ID","IGHV.status","TP53","trisomy12","del17p")], by = c(patientID = "Patient.ID")) %>%
mutate(TP53 = as.numeric(as.character(TP53)),
del17p = as.numeric(as.character(del17p))) %>%
mutate(`TP53.del17p` = as.numeric(TP53 | del17p),
IGHV = factor(ifelse(IGHV.status %in% "U",1,0))) %>%
select(-TP53, -del17p,-IGHV.status) %>%
mutate(age = age/10) Multi-variate test
cTab.ttt <- lapply(filter(uniRes, outcome == "TTT", p.adj <=0.1)$id, function(n) {
risk0 <- riskTab
expr <- protMat[n,]
expr <- (expr - mean(expr,na.rm=TRUE))/sd(expr,na.rm = TRUE)
risk1 <- riskTab %>% mutate(protExpr = expr[patientID])
res0 <- summary(runCox(survTab, risk0, "TTT","treatedAfter"))
fullModel <- runCox(survTab, risk1, "TTT","treatedAfter")
res1 <- summary(fullModel)
tibble(id = n, c0 = res0$concordance[1], c1 = res1$concordance[1],
se0 = res0$concordance[2],se1 = res1$concordance[2],
ci0 = se0*1.96, ci1 = se1*1.96,
p = res1$coefficients["protExpr",5],
fullModel = list(fullModel))
}) %>% bind_rows() %>% mutate(diffC = c1-c0) %>%
arrange(desc(diffC)) %>%
mutate(name=rowData(protCLL[id,])$hgnc_symbol,
outcome = "TTT")
cTab.os <- lapply(filter(uniRes, outcome == "OS", p.adj<=0.1)$id, function(n) {
risk0 <- riskTab
expr <- protMat[n,]
expr <- (expr - mean(expr,na.rm=TRUE))/sd(expr,na.rm = TRUE)
risk1 <- riskTab %>% mutate(protExpr = expr[patientID])
res0 <- summary(runCox(survTab, risk0, "OS","died"))
fullModel <- runCox(survTab, risk1, "OS","died")
res1 <- summary(fullModel)
tibble(id = n, c0 = res0$concordance[1], c1 = res1$concordance[1],
se0 = res0$concordance[2],se1 = res1$concordance[2],
ci0 = se0*1.96, ci1 = se1*1.96,
p = res1$coefficients["protExpr",5],
fullModel = list(fullModel))
}) %>% bind_rows() %>% mutate(diffC = c1-c0) %>%
arrange(desc(diffC)) %>%
mutate(name=rowData(protCLL[id,])$hgnc_symbol,
outcome = "OS")
cTab <- bind_rows(cTab.ttt, cTab.os) %>%
mutate(p.adj = p.adjust(p, method = "BH")) %>%
arrange(p)A table showing idependent protein markers
cTab %>% filter(p.adj <= 0.05) %>% mutate_if(is.numeric, formatC, digits=2,format="e") %>%
select(name, p, p.adj, outcome) %>% DT::datatable()Forest plot of selected markers
plotHazard <- function(survRes, protName, title = "") {
sumTab <- summary(survRes)$coefficients
confTab <- summary(survRes)$conf.int
#correct feature name
nameOri <- rownames(sumTab)
nameMod <- substr(nameOri, 1, nchar(nameOri) -1)
plotTab <- tibble(feature = rownames(sumTab),
nameMod = substr(nameOri, 1, nchar(nameOri) -1),
HR = sumTab[,2],
p = sumTab[,5],
Upper = confTab[,4],
Lower = confTab[,3]) %>%
mutate(feature = ifelse(nameMod %in% names(survRes$xlevels), nameMod, feature)) %>%
mutate(feature = str_replace(feature, "[.]","/")) %>%
mutate(feature = str_replace(feature, "[_]","-")) %>%
mutate(candidate = ifelse(feature == "protExpr", "yes","no")) %>%
mutate(feature = ifelse(feature == "protExpr", protName, feature)) %>%
arrange(desc(abs(p))) %>% mutate(feature = factor(feature, levels = feature)) #%>%
#mutate(type = ifelse(HR >1 ,"up","down")) %>%
# mutate(Upper = ifelse(Upper > 10, 10, Upper))
p <- ggplot(plotTab, aes(x=feature, y = HR, color = candidate)) +
geom_hline(yintercept = 1, linetype = "dotted") +
geom_point(position = position_dodge(width=0.8), size=3) +
geom_errorbar(aes(ymin = Lower, ymax = Upper), width = 0.3, size=1) +
geom_text(position = position_nudge(x = 0.3),
aes(y = HR, label = sprintf("italic(P)~'='~'%s'",
formatNum(p, digits = 1))),
color = "black", size =5, parse = TRUE) +
#expand_limits(y=c(-0.5,0))+
scale_color_manual(values = c(yes = "darkred", no = "black")) +
ggtitle(title) + scale_y_log10() +
ylab("Hazard ratio") +
coord_flip() +
theme_full +
theme(legend.position = "none", axis.title.y = element_blank())
return(p)
}NFKB2
protName <- "NFKB2"
outcomeName <- "TTT"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.prmt5 <- plotHazard(survRes, protName, outcomeName)
hr.prmt5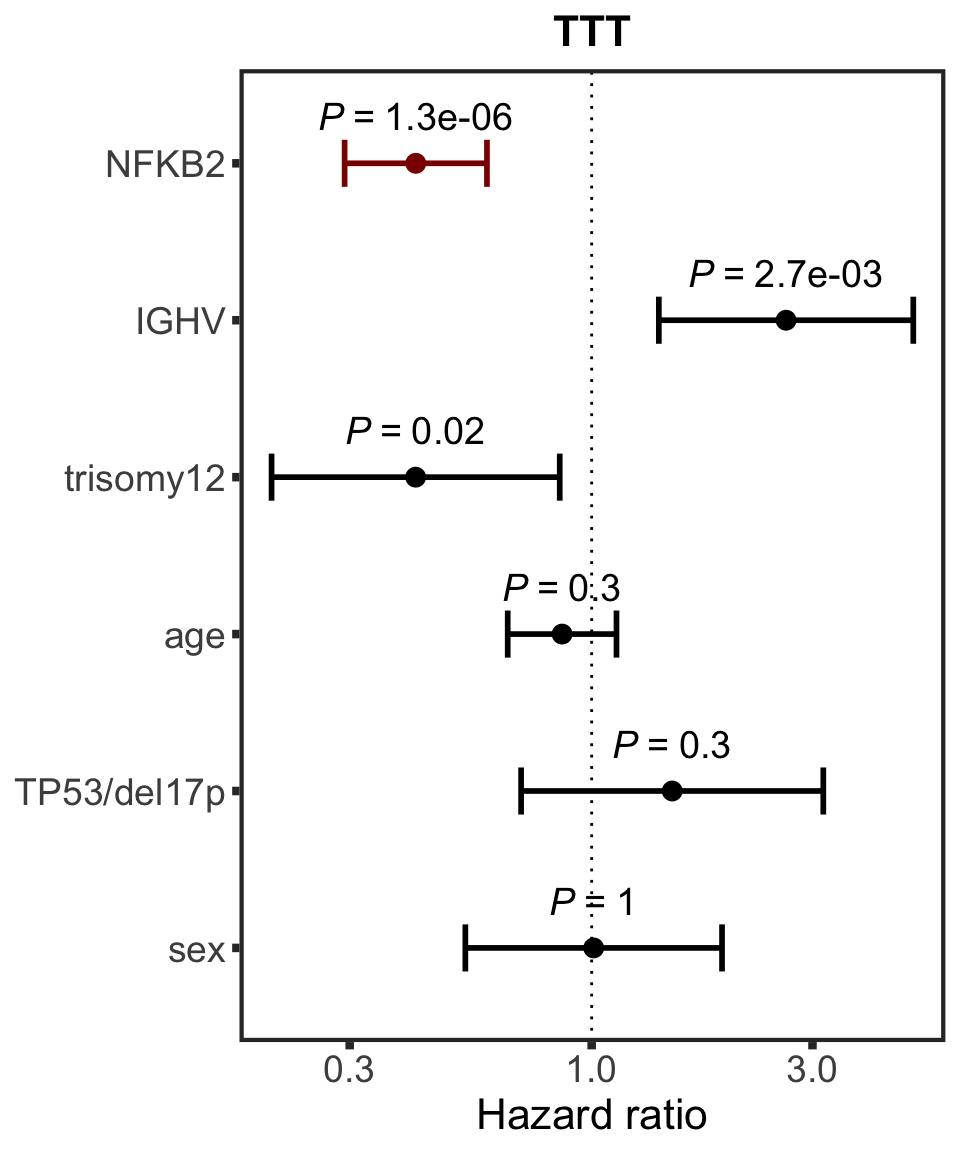
PURA
protName <- "PURA"
outcomeName <- "TTT"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.prmt5 <- plotHazard(survRes, protName, outcomeName)
hr.prmt5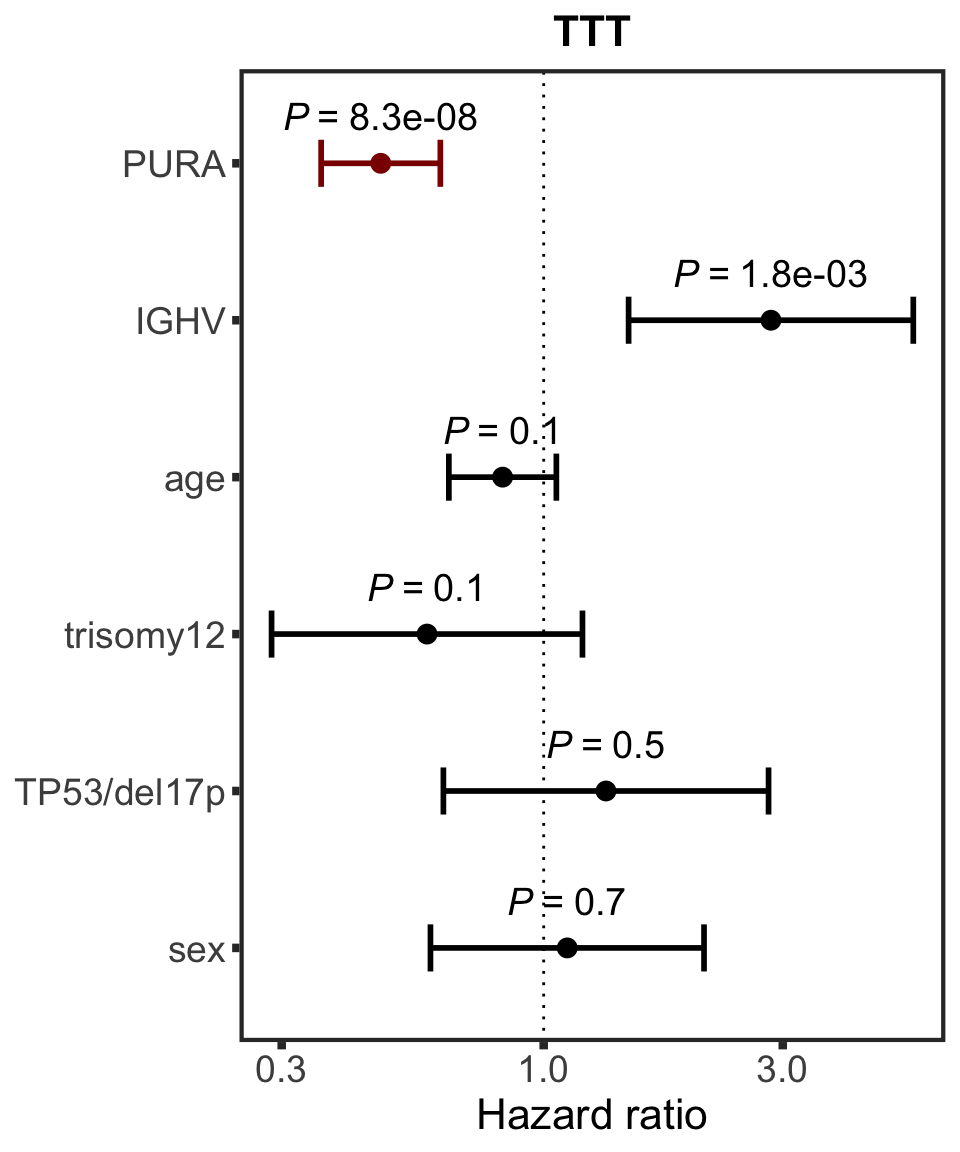
PRMT5
protName <- "PRMT5"
outcomeName <- "TTT"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.prmt5 <- plotHazard(survRes, protName, outcomeName)
hr.prmt5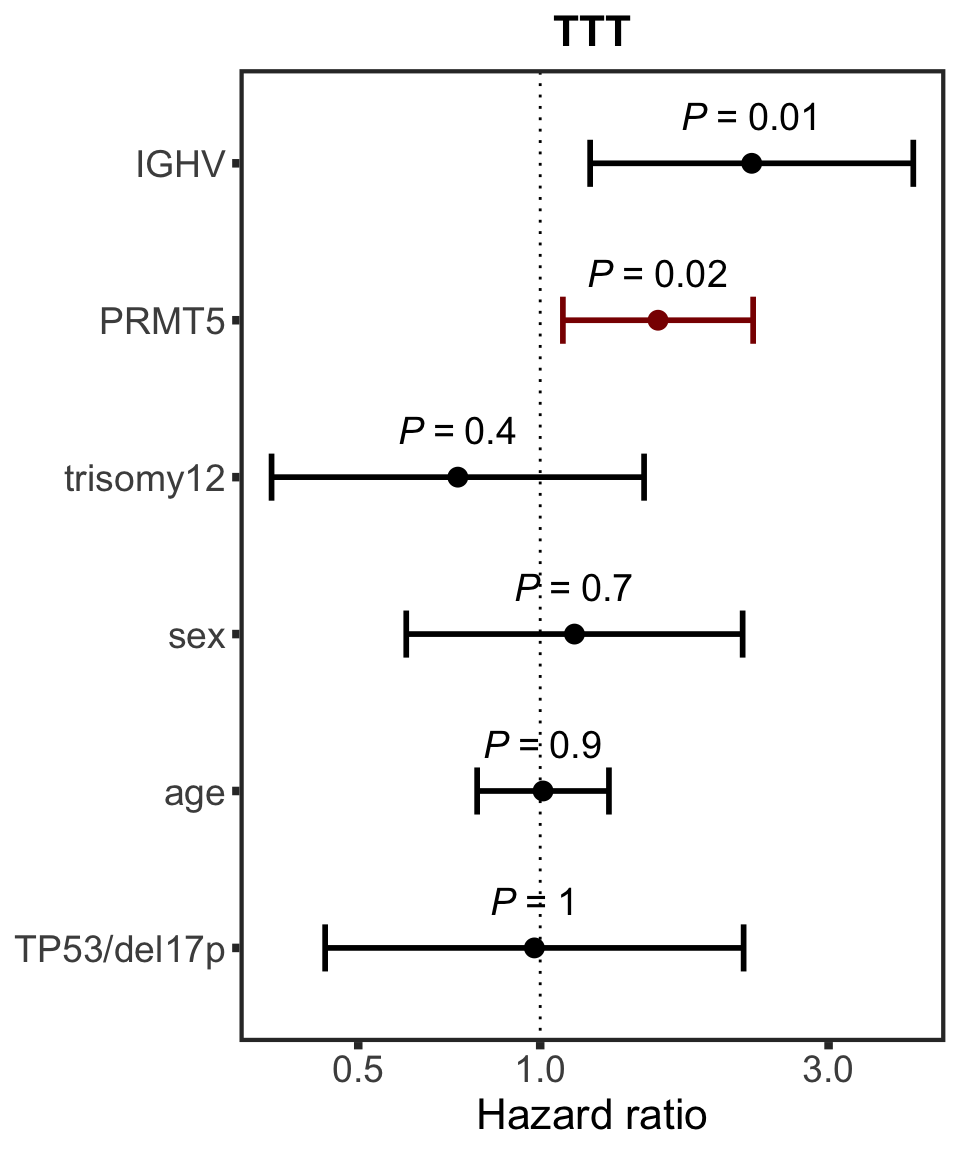
PYGB
protName <- "PYGB"
outcomeName <- "TTT"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.pygb <- plotHazard(survRes, protName, outcomeName)
hr.pygb
PES1
protName <- "PES1"
outcomeName <- "TTT"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.pes1 <- plotHazard(survRes, protName, outcomeName)
hr.pes1
RRAS2
protName <- "RRAS2"
outcomeName <- "OS"
survRes <- filter(cTab, outcome == outcomeName , name == protName)$fullModel[[1]]
hr.rras <- plotHazard(survRes, protName, outcomeName)
hr.rras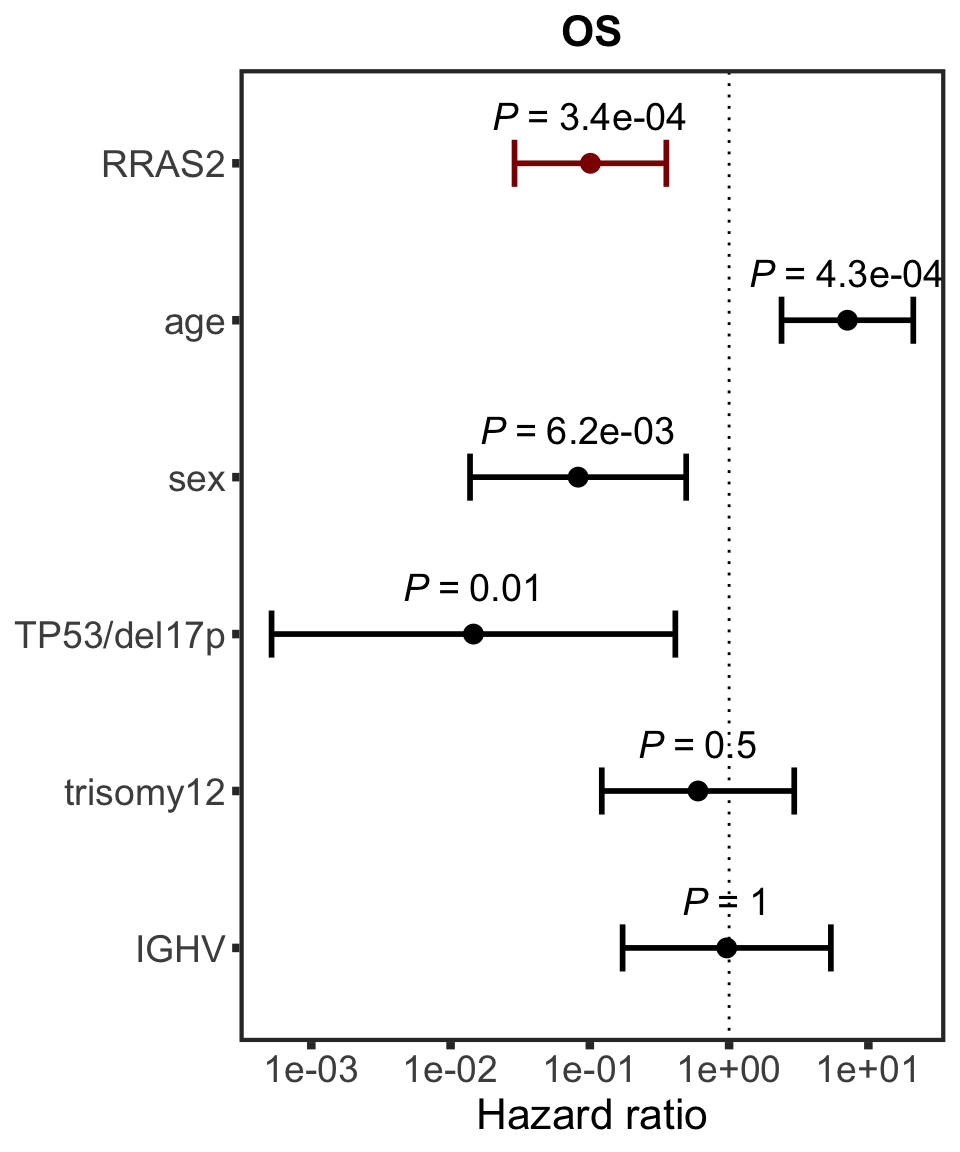
Bar plots comparing C-index
TTT
nameList <- c("PRMT5","PYGB","PES1")
cTab.sub <- filter(cTab, outcome == "TTT", name %in% nameList)
plotList <- lapply(seq(nrow(cTab.sub)), function(i) {
plotTab <- tibble(model = c("knownRisks","plusProtein"),
cindex = c(cTab.sub[i,]$c0, cTab.sub[i,]$c1),
ci = c(cTab.sub[i,]$ci0,cTab.sub[i,]$ci1))
ggplot(plotTab, aes(x=model, y=cindex, fill = model)) +
geom_bar(stat="identity",width=0.6) +
geom_errorbar(aes(ymin=cindex + ci, ymax = cindex-ci), width=0.5) +
theme_full+ scale_fill_manual(values = colList[6:7]) +
ggtitle(cTab.ttt[i,]$name) + theme(legend.position = "none") +
ylab("Harrell's C-index")
})
cTTT <- plot_grid(plotlist = plotList, ncol =3)
cTTT
TTT (other candiates)
nameList <- c("PURA","NFKB2","CLTB")
cTab.sub <- filter(cTab, outcome == "TTT", name %in% nameList)
plotList <- lapply(seq(nrow(cTab.sub)), function(i) {
plotTab <- tibble(model = c("knownRisks","plusProtein"),
cindex = c(cTab.sub[i,]$c0, cTab.sub[i,]$c1),
ci = c(cTab.sub[i,]$ci0,cTab.sub[i,]$ci1))
ggplot(plotTab, aes(x=model, y=cindex, fill = model)) +
geom_bar(stat="identity",width=0.6) +
geom_errorbar(aes(ymin=cindex + ci, ymax = cindex-ci), width=0.5) +
theme_full+ scale_fill_manual(values = colList[6:7]) +
ggtitle(cTab.ttt[i,]$name) + theme(legend.position = "none") +
ylab("Harrell's C-index")
})
cTTT <- plot_grid(plotlist = plotList, ncol =3)
cTTT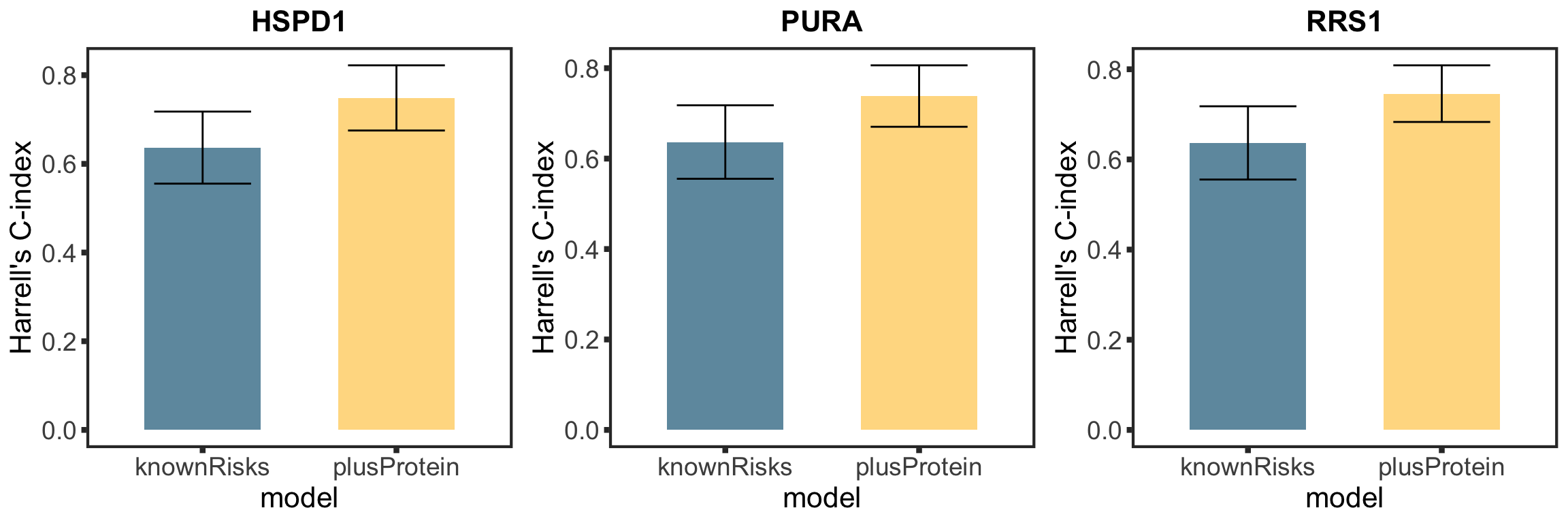
Save for comparison
save(uniRes, cTab, file ="../output/resOutcome_batch13.RData")
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] BiocParallel_1.22.0 latex2exp_0.4.0
[3] forcats_0.5.1 stringr_1.4.0
[5] dplyr_1.0.5 purrr_0.3.4
[7] readr_1.4.0 tidyr_1.1.3
[9] tibble_3.1.0 tidyverse_1.3.0
[11] cowplot_1.1.1 SummarizedExperiment_1.18.2
[13] DelayedArray_0.14.1 matrixStats_0.58.0
[15] Biobase_2.48.0 GenomicRanges_1.40.0
[17] GenomeInfoDb_1.24.2 IRanges_2.22.2
[19] S4Vectors_0.26.1 BiocGenerics_0.34.0
[21] glmnet_4.1-1 Matrix_1.3-2
[23] maxstat_0.7-25 survminer_0.4.9
[25] ggpubr_0.4.0 ggplot2_3.3.3
[27] survival_3.2-7 jyluMisc_0.1.5
[29] pheatmap_1.0.12 limma_3.44.3
loaded via a namespace (and not attached):
[1] readxl_1.3.1 backports_1.2.1 fastmatch_1.1-0
[4] drc_3.0-1 workflowr_1.6.2 igraph_1.2.6
[7] shinydashboard_0.7.1 splines_4.0.2 crosstalk_1.1.1
[10] TH.data_1.0-10 digest_0.6.27 foreach_1.5.1
[13] htmltools_0.5.1.1 fansi_0.4.2 magrittr_2.0.1
[16] cluster_2.1.1 openxlsx_4.2.3 modelr_0.1.8
[19] sandwich_3.0-0 piano_2.4.0 colorspace_2.0-0
[22] rvest_1.0.0 haven_2.3.1 xfun_0.21
[25] crayon_1.4.1 RCurl_1.98-1.2 jsonlite_1.7.2
[28] zoo_1.8-9 iterators_1.0.13 glue_1.4.2
[31] gtable_0.3.0 zlibbioc_1.34.0 XVector_0.28.0
[34] car_3.0-10 shape_1.4.5 abind_1.4-5
[37] scales_1.1.1 mvtnorm_1.1-1 DBI_1.1.1
[40] relations_0.6-9 rstatix_0.7.0 Rcpp_1.0.6
[43] plotrix_3.8-1 xtable_1.8-4 foreign_0.8-81
[46] km.ci_0.5-2 DT_0.17 htmlwidgets_1.5.3
[49] httr_1.4.2 fgsea_1.14.0 gplots_3.1.1
[52] RColorBrewer_1.1-2 ellipsis_0.3.1 farver_2.1.0
[55] pkgconfig_2.0.3 sass_0.3.1 dbplyr_2.1.0
[58] utf8_1.1.4 labeling_0.4.2 tidyselect_1.1.0
[61] rlang_0.4.10 later_1.1.0.1 munsell_0.5.0
[64] cellranger_1.1.0 tools_4.0.2 visNetwork_2.0.9
[67] cli_2.3.1 generics_0.1.0 broom_0.7.5
[70] evaluate_0.14 fastmap_1.1.0 yaml_2.2.1
[73] knitr_1.31 fs_1.5.0 zip_2.1.1
[76] survMisc_0.5.5 caTools_1.18.1 nlme_3.1-152
[79] mime_0.10 slam_0.1-48 xml2_1.3.2
[82] rstudioapi_0.13 compiler_4.0.2 curl_4.3
[85] ggsignif_0.6.1 marray_1.66.0 reprex_1.0.0
[88] bslib_0.2.4 stringi_1.5.3 highr_0.8
[91] lattice_0.20-41 shinyjs_2.0.0 KMsurv_0.1-5
[94] vctrs_0.3.6 pillar_1.5.1 lifecycle_1.0.0
[97] jquerylib_0.1.3 data.table_1.14.0 bitops_1.0-6
[100] httpuv_1.5.5 R6_2.5.0 promises_1.2.0.1
[103] KernSmooth_2.23-18 gridExtra_2.3 rio_0.5.26
[106] codetools_0.2-18 MASS_7.3-53.1 gtools_3.8.2
[109] exactRankTests_0.8-31 assertthat_0.2.1 rprojroot_2.0.2
[112] withr_2.4.1 multcomp_1.4-16 GenomeInfoDbData_1.2.3
[115] mgcv_1.8-34 hms_1.0.0 grid_4.0.2
[118] rmarkdown_2.7 carData_3.0-4 git2r_0.28.0
[121] sets_1.0-18 shiny_1.6.0 lubridate_1.7.10