Section 2: an overview of the associations between genomics and protein expressions
Junyan Lu
2020-06-23
Last updated: 2020-08-06
Checks: 5 2
Knit directory: Proteomics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200227) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- unnamed-chunk-15
- unnamed-chunk-17
- unnamed-chunk-6
- unnamed-chunk-8
To ensure reproducibility of the results, delete the cache directory manuscript_S2_genomicAssociation_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3fb50c5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/analysisDrugResponses_IC50_cache/
Ignored: analysis/analysisDrugResponses_cache/
Ignored: analysis/complexAnalysis_IGHV_alternative_cache/
Ignored: analysis/complexAnalysis_IGHV_cache/
Ignored: analysis/complexAnalysis_trisomy12_alteredPQR_cache/
Ignored: analysis/complexAnalysis_trisomy12_alternative_cache/
Ignored: analysis/complexAnalysis_trisomy12_cache/
Ignored: analysis/correlateCLLPD_cache/
Ignored: analysis/correlateRNAexpression_cache/
Ignored: analysis/manuscript_S1_Overview_cache/
Ignored: analysis/manuscript_S2_genomicAssociation_cache/
Ignored: analysis/manuscript_S3_trisomy12_cache/
Ignored: analysis/manuscript_S4_IGHV_cache/
Ignored: analysis/manuscript_S5_trisomy19_cache/
Ignored: analysis/manuscript_S6_del11q_cache/
Ignored: analysis/manuscript_S7_SF3B1_cache/
Ignored: analysis/manuscript_S8_drugResponse_Outcomes_cache/
Ignored: analysis/predictOutcome_cache/
Ignored: code/.Rhistory
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/.trisomy12_norm.pdf
Untracked: analysis/CNVanalysis_11q.Rmd
Untracked: analysis/CNVanalysis_trisomy12.Rmd
Untracked: analysis/CNVanalysis_trisomy19.Rmd
Untracked: analysis/SUGP1_splicing.svg.xml
Untracked: analysis/analysisDrugResponses.Rmd
Untracked: analysis/analysisDrugResponses_IC50.Rmd
Untracked: analysis/analysisPCA.Rmd
Untracked: analysis/analysisPreliminary_LUMOS.Rmd
Untracked: analysis/analysisPreliminary_timsTOF_Hela.Rmd
Untracked: analysis/analysisPreliminary_timsTOF_new.Rmd
Untracked: analysis/analysisSplicing.Rmd
Untracked: analysis/analysisTrisomy19.Rmd
Untracked: analysis/annotateCNV.Rmd
Untracked: analysis/comparePlatforms_LUMOS_helaTimsTOF.Rmd
Untracked: analysis/comparePlatforms_LUMOS_newTimsTOF.Rmd
Untracked: analysis/comparePlatforms_newTimsTOF_helaTimsTOF copy.Rmd
Untracked: analysis/complexAnalysis_IGHV.Rmd
Untracked: analysis/complexAnalysis_IGHV_alternative.Rmd
Untracked: analysis/complexAnalysis_overall.Rmd
Untracked: analysis/complexAnalysis_trisomy12.Rmd
Untracked: analysis/complexAnalysis_trisomy12_alternative.Rmd
Untracked: analysis/correlateGenomic_PC12adjusted.Rmd
Untracked: analysis/correlateGenomic_noBlock.Rmd
Untracked: analysis/correlateGenomic_noBlock_MCLL.Rmd
Untracked: analysis/correlateGenomic_noBlock_UCLL.Rmd
Untracked: analysis/correlateRNAexpression.Rmd
Untracked: analysis/default.css
Untracked: analysis/del11q.pdf
Untracked: analysis/del11q_norm.pdf
Untracked: analysis/manuscript_S0_PrepareData.Rmd
Untracked: analysis/manuscript_S1_Overview.Rmd
Untracked: analysis/manuscript_S2_genomicAssociation.Rmd
Untracked: analysis/manuscript_S3_trisomy12.Rmd
Untracked: analysis/manuscript_S4_IGHV.Rmd
Untracked: analysis/manuscript_S5_trisomy19.Rmd
Untracked: analysis/manuscript_S6_del11q.Rmd
Untracked: analysis/manuscript_S7_SF3B1.Rmd
Untracked: analysis/manuscript_S8_drugResponse_Outcomes.Rmd
Untracked: analysis/peptideValidate.Rmd
Untracked: analysis/plotExpressionCNV.Rmd
Untracked: analysis/processPeptides_LUMOS.Rmd
Untracked: analysis/processProteomics_timsTOF_Hela.Rmd
Untracked: analysis/processProteomics_timsTOF_new.Rmd
Untracked: analysis/qualityControl_timsTOF_Hela.Rmd
Untracked: analysis/qualityControl_timsTOF_new.Rmd
Untracked: analysis/style.css
Untracked: analysis/test.pdf
Untracked: analysis/test.svg
Untracked: analysis/tri12Enrich.pdf
Untracked: analysis/trisomy12.pdf
Untracked: analysis/trisomy12_AFcor.Rmd
Untracked: analysis/trisomy12_norm.pdf
Untracked: code/AlteredPQR.R
Untracked: code/utils.R
Untracked: data/190909_CLL_prot_abund_med_norm.tsv
Untracked: data/190909_CLL_prot_abund_no_norm.tsv
Untracked: data/200725_cll_diaPASEF_direct_reports/
Untracked: data/200728_cll_diaPASEF_direct_plus_hela_reports/
Untracked: data/20190423_Proteom_submitted_samples_bereinigt.xlsx
Untracked: data/20191025_Proteom_submitted_samples_final.xlsx
Untracked: data/LUMOS/
Untracked: data/LUMOS_peptides/
Untracked: data/LUMOS_protAnnotation.csv
Untracked: data/LUMOS_protAnnotation_fix.csv
Untracked: data/SampleAnnotation_cleaned.xlsx
Untracked: data/example_proteomics_data
Untracked: data/facTab_IC50atLeast3New.RData
Untracked: data/gmts/
Untracked: data/mapEnsemble.txt
Untracked: data/mapSymbol.txt
Untracked: data/proteins_in_complexes
Untracked: data/pyprophet_export_aligned.csv
Untracked: data/timsTOF_protAnnotation.csv
Untracked: output/Fig1A.pdf
Untracked: output/Fig1A.png
Untracked: output/Fig1A.pptx
Untracked: output/LUMOS_processed.RData
Untracked: output/MSH6_splicing.svg
Untracked: output/SUGP1_splicing.eps
Untracked: output/SUGP1_splicing.pdf
Untracked: output/SUGP1_splicing.svg
Untracked: output/cnv_plots.zip
Untracked: output/cnv_plots/
Untracked: output/cnv_plots_norm.zip
Untracked: output/ddsrna_enc.RData
Untracked: output/deResList.RData
Untracked: output/deResList_timsTOF.RData
Untracked: output/dxdCLL.RData
Untracked: output/dxdCLL2.RData
Untracked: output/encMap.RData
Untracked: output/exprCNV.RData
Untracked: output/exprCNV_enc.RData
Untracked: output/lassoResults_CPS.RData
Untracked: output/lassoResults_IC50.RData
Untracked: output/patMeta_enc.RData
Untracked: output/pepCLL_lumos.RData
Untracked: output/pepCLL_lumos_enc.RData
Untracked: output/pepTab_lumos.RData
Untracked: output/pheno1000_enc.RData
Untracked: output/pheno1000_main.RData
Untracked: output/plotCNV_allChr11_diff.pdf
Untracked: output/plotCNV_del11q_sum.pdf
Untracked: output/proteomic_LUMOS_20200227.RData
Untracked: output/proteomic_LUMOS_20200320.RData
Untracked: output/proteomic_LUMOS_20200430.RData
Untracked: output/proteomic_LUMOS_enc.RData
Untracked: output/proteomic_timsTOF_20200227.RData
Untracked: output/proteomic_timsTOF_Hela_20200806.RData
Untracked: output/proteomic_timsTOF_enc.RData
Untracked: output/proteomic_timsTOF_new_20200806.RData
Untracked: output/splicingResults.RData
Untracked: output/survival_enc.RData
Untracked: output/timsTOF_processed.RData
Untracked: plotCNV_del11q_diff.pdf
Untracked: supp_latex/
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/analysisSF3B1.Rmd
Modified: analysis/compareProteomicsRNAseq.Rmd
Modified: analysis/correlateCLLPD.Rmd
Modified: analysis/correlateGenomic.Rmd
Deleted: analysis/correlateGenomic_removePC.Rmd
Modified: analysis/correlateMIR.Rmd
Modified: analysis/correlateMethylationCluster.Rmd
Modified: analysis/index.Rmd
Modified: analysis/predictOutcome.Rmd
Modified: analysis/processProteomics_LUMOS.Rmd
Modified: analysis/qualityControl_LUMOS.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Preprocessing
Process proteomics data
protCLL <- protCLL[rowData(protCLL)$uniqueMap,]
protMat <- assays(protCLL)[["count"]] #without imputationPrepare genomic background
Get Mutations with at least 5 cases
geneMat <- patMeta[match(colnames(protMat), patMeta$Patient.ID),] %>%
select(Patient.ID, IGHV.status, del11p:U1) %>%
mutate_if(is.factor, as.character) %>% mutate(IGHV.status = ifelse(IGHV.status == "M", 1,0)) %>%
mutate_at(vars(-Patient.ID), as.numeric) %>% #assign a few unknown mutated cases to wildtype
data.frame() %>% column_to_rownames("Patient.ID")
geneMat <- geneMat[,apply(geneMat,2, function(x) sum(x %in% 1, na.rm = TRUE))>=5]Mutations that will be tested
colnames(geneMat) [1] "IGHV.status" "del11q" "del13q" "trisomy12" "trisomy19"
[6] "DDX3X" "EGR2" "NOTCH1" "SF3B1" "TP53" Plot to summarise genomic background
sortTab <- function(sumTab) {
i <- ncol(sumTab)
#print(i)
if (i == 1) {
#print(rownames(sumTab)[order(sumTab[,i])])
return(rownames(sumTab)[order(sumTab[,i])])
}
orderRow <- c(sortTab(sumTab[sumTab[,i]==0, seq(1,i-1), drop = FALSE]), sortTab(sumTab[sumTab[,i]==1, seq(1,i-1), drop = FALSE]))
return(orderRow)
}
geneMat.sort <- geneMat
geneMat.sort[is.na(geneMat.sort)] <- 0
sortedGene <- names(sort(colSums(geneMat.sort)))
geneMat.sort <- geneMat.sort[,sortedGene]
sortedPat <- sortTab(geneMat.sort)plotTab <- geneMat %>% rownames_to_column("patID") %>% mutate_all(as.character) %>%
gather(key = "gene", value = "status", -patID) %>%
mutate(gene = factor(gene, levels = sortedGene), patID = factor(patID, levels = sortedPat)) %>%
mutate(status = ifelse(is.na(status),"NA",status))
ggplot(plotTab, aes(x=patID, y = gene, fill = status)) +
geom_tile(color = "black") +
theme_minimal() +
scale_fill_manual(values = c("1" = colList[4],"0" ="white", "NA" = "grey80")) +
theme(axis.text.x = element_text(angle = 90, hjust =1, vjust=0.5),
panel.grid = element_blank(), legend.position = "none") +
ylab("") + xlab("Patient ID")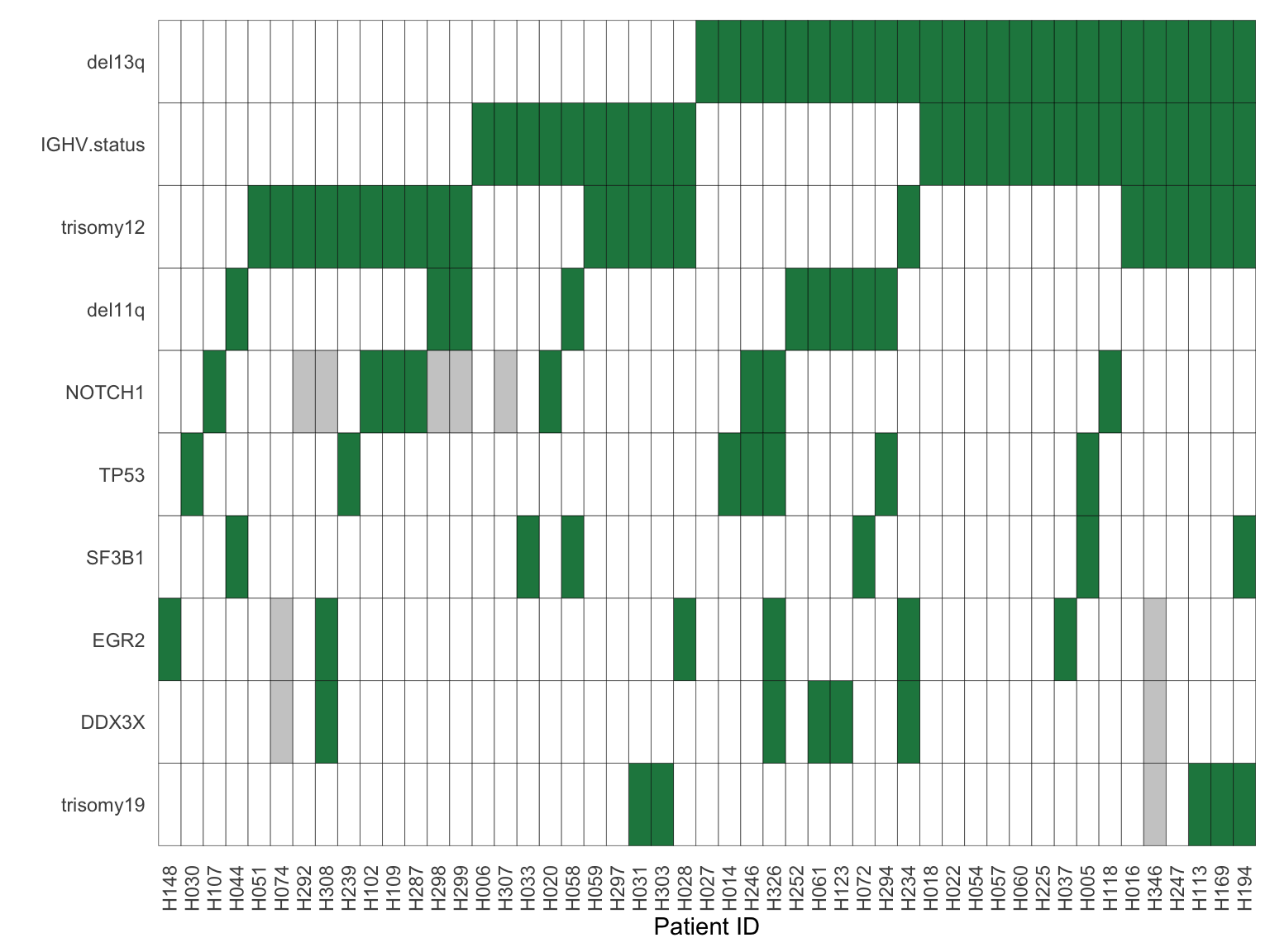
Association test using proDA
For IGHV and trisomy12
Fit the probailistic dropout model
designMat <- geneMat[ ,c("IGHV.status","trisomy12")]
fit <- proDA(protMat, design = ~ .,
col_data = designMat)Test for differentially expressed proteins
resList.ighvTri12 <- lapply(c("IGHV.status","trisomy12"), function(n) {
contra <- n
resTab <- test_diff(fit, contra) %>%
dplyr::rename(id = name, logFC = diff, t=t_statistic,
P.Value = pval, adj.P.Val = adj_pval) %>%
mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
select(name, id, logFC, t, P.Value, adj.P.Val) %>%
arrange(P.Value) %>% mutate(Gene = n) %>%
as_tibble()
}) %>% bind_rows()Test for other variantions (blocking for IGHV and trisomy12)
Fit the probailistic dropout model and test for differentially expressed proteins
otherGenes <- colnames(geneMat)[!colnames(geneMat)%in% c("IGHV.status","trisomy12")]
resList <- lapply(otherGenes, function(n) {
designMat <- geneMat[,c("IGHV.status","trisomy12",n)]
designMat <- designMat[!is.na(designMat[[n]]),]
testMat <- protMat[,rownames(designMat)]
fit <- proDA(testMat, design = ~ .,
col_data = designMat)
contra <- n
resTab <- test_diff(fit, contra) %>%
dplyr::rename(id = name, logFC = diff, t=t_statistic,
P.Value = pval, adj.P.Val = adj_pval) %>%
mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
select(name, id, logFC, t, P.Value, adj.P.Val) %>%
arrange(P.Value) %>% mutate(Gene = n) %>%
as_tibble()
resTab
}) %>% bind_rows()Combine the results
resList <- bind_rows(resList.ighvTri12, resList)
#Adjusting p values
#using BH
resList <- mutate(resList, adj.P.global = p.adjust(P.Value, method = "BH"))
#using IHW
ihwRes <- ihw(P.Value ~ factor(Gene), data= resList, alpha=0.1)
resList <- mutate(resList, adj.P.IHW = adj_pvalues(ihwRes))save(resList, file = "../output/deResList.RData")load("../output/deResList.RData")Summarise significant associations
Bar plot of number of significant associations (10% FDR)
plotTab <- resList %>% group_by(Gene) %>%
summarise(nFDR.local = sum(adj.P.Val <= 0.1),
nFDR.global = sum(adj.P.global <= 0.1),
nFDR.IHW = sum(adj.P.IHW <= 0.1),
nP = sum(P.Value < 0.05))
#use IHW for adjusting p-values
resList <- mutate(resList, adj.P.Val = adj.P.IHW)plotTab <- arrange(plotTab, desc(nFDR.IHW)) %>% mutate(Gene = factor(Gene, levels = Gene))
ggplot(plotTab, aes(x=Gene, y = nFDR.IHW)) + geom_bar(stat="identity",fill=colList[2]) +
geom_text(aes(label = paste0("n=", nFDR.IHW)),vjust=-1,col=colList[1]) + ylim(0,1200) +
theme_half + theme(axis.text.x = element_text(angle = 90, hjust=1, vjust=0.5)) +
ylab("Number of associations\n(10% FDR)") + xlab("")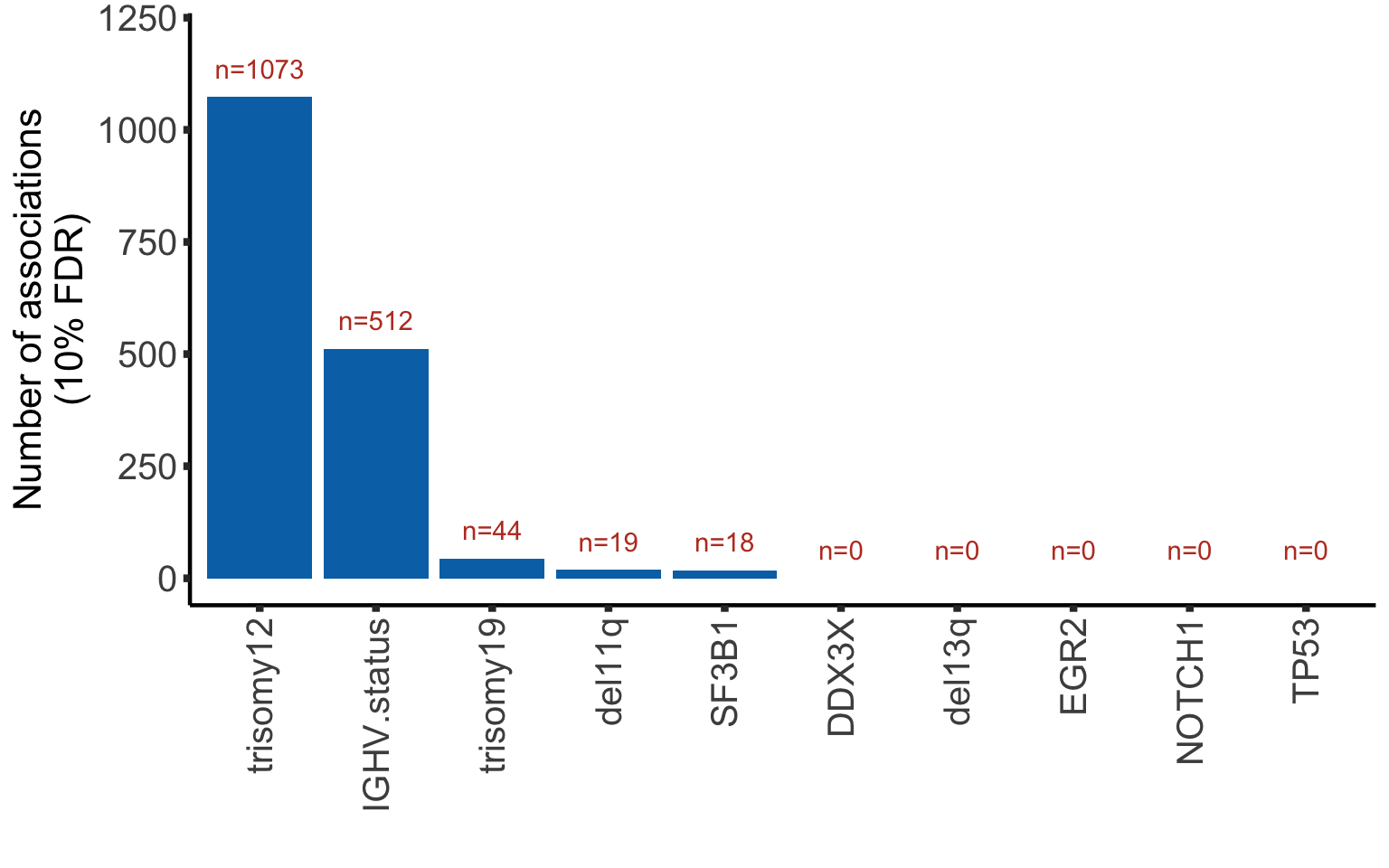
Bar plot of number of raw P-value < 0.05
plotTab <- arrange(plotTab, desc(nP)) %>% mutate(Gene = factor(Gene, levels = Gene))
ggplot(plotTab, aes(x=Gene, y = nP)) + geom_bar(stat="identity",fill=colList[2]) +
geom_text(aes(label = paste0("n=", nP)),vjust=-1,col=colList[1]) + ylim(0,1500) +
theme_half + theme(axis.text.x = element_text(angle = 90, hjust=1, vjust=0.5)) +
ylab("Number of associations\n(nominal P-value < 0.05)") + xlab("")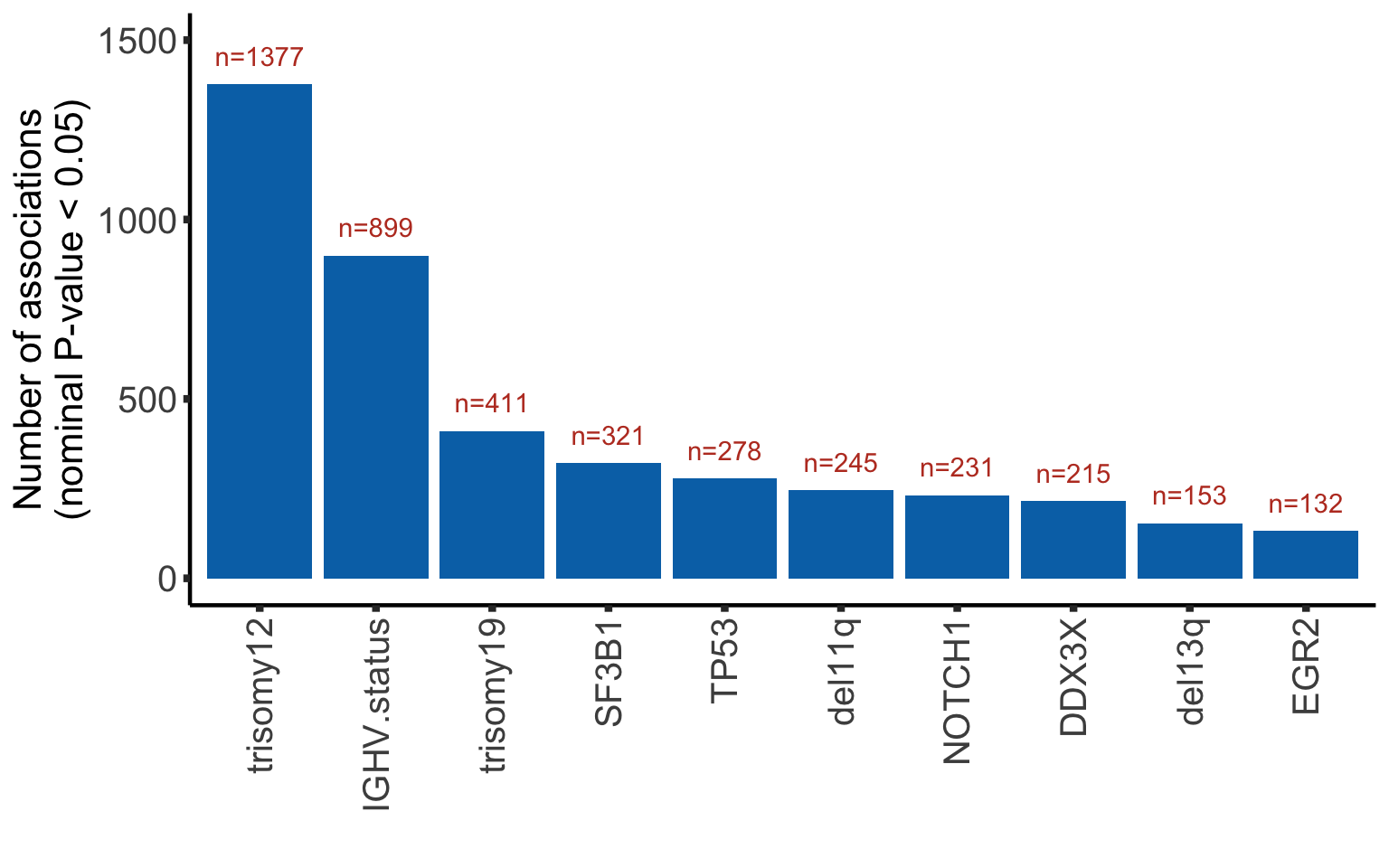
Association test in timsTOF data
For IGHV and trisomy12
Load timsTOF data
load("../output/proteomic_timsTOF_enc.RData")
protMat <- assays(protCLL)[["count"]] #without imputationGenetic data
geneMat <- patMeta[match(colnames(protMat), patMeta$Patient.ID),] %>%
select(Patient.ID, IGHV.status, trisomy12, SF3B1, trisomy19, del11q) %>%
mutate_if(is.factor, as.character) %>% mutate(IGHV.status = ifelse(IGHV.status == "M", 1,0)) %>%
mutate_at(vars(-Patient.ID), as.numeric) %>% #assign a few unknown mutated cases to wildtype
data.frame() %>% column_to_rownames("Patient.ID")Fit the probailistic dropout model
designMat <- geneMat[ ,c("IGHV.status","trisomy12")]
fit <- proDA(protMat, design = ~ .,
col_data = designMat)Test for differentially expressed proteins
resList.ighvTri12 <- lapply(c("IGHV.status","trisomy12"), function(n) {
contra <- n
resTab <- test_diff(fit, contra) %>%
dplyr::rename(id = name, logFC = diff, t=t_statistic,
P.Value = pval, adj.P.Val = adj_pval) %>%
mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
select(name, id, logFC, t, P.Value, adj.P.Val) %>%
arrange(P.Value) %>% mutate(Gene = n) %>%
as_tibble()
}) %>% bind_rows()Test for other variantions (blocking for IGHV and trisomy12)
Fit the probailistic dropout model and test for differentially expressed proteins
otherGenes <- colnames(geneMat)[!colnames(geneMat)%in% c("IGHV.status","trisomy12")]
resList <- lapply(otherGenes, function(n) {
designMat <- geneMat[,c("IGHV.status","trisomy12",n)]
designMat <- designMat[!is.na(designMat[[n]]),]
testMat <- protMat[,rownames(designMat)]
fit <- proDA(testMat, design = ~ .,
col_data = designMat)
contra <- n
resTab <- test_diff(fit, contra) %>%
dplyr::rename(id = name, logFC = diff, t=t_statistic,
P.Value = pval, adj.P.Val = adj_pval) %>%
mutate(name = rowData(protCLL[id,])$hgnc_symbol) %>%
select(name, id, logFC, t, P.Value, adj.P.Val) %>%
arrange(P.Value) %>% mutate(Gene = n) %>%
as_tibble()
resTab
}) %>% bind_rows()Combine the results
resList <- bind_rows(resList.ighvTri12, resList)
#Adjusting p values
#using BH
resList <- mutate(resList, adj.P.global = p.adjust(P.Value, method = "BH"))
#using IHW
ihwRes <- ihw(P.Value ~ factor(Gene), data= resList, alpha=0.1)
resList <- mutate(resList, adj.P.IHW = adj_pvalues(ihwRes))save(resList, file = "../output/deResList_timsTOF.RData")
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] latex2exp_0.4.0 forcats_0.5.0
[3] stringr_1.4.0 dplyr_1.0.0
[5] purrr_0.3.4 readr_1.3.1
[7] tidyr_1.1.0 tibble_3.0.3
[9] ggplot2_3.3.2 tidyverse_1.3.0
[11] IHW_1.14.0 proDA_1.1.2
[13] DESeq2_1.26.0 SummarizedExperiment_1.16.1
[15] DelayedArray_0.12.3 BiocParallel_1.20.1
[17] matrixStats_0.56.0 Biobase_2.46.0
[19] GenomicRanges_1.38.0 GenomeInfoDb_1.22.1
[21] IRanges_2.20.2 S4Vectors_0.24.4
[23] BiocGenerics_0.32.0 limma_3.42.2
loaded via a namespace (and not attached):
[1] colorspace_1.4-1 ellipsis_0.3.1 rprojroot_1.3-2
[4] htmlTable_2.0.1 XVector_0.26.0 base64enc_0.1-3
[7] fs_1.4.2 rstudioapi_0.11 farver_2.0.3
[10] bit64_0.9-7 fansi_0.4.1 AnnotationDbi_1.48.0
[13] lubridate_1.7.9 xml2_1.3.2 splines_3.6.0
[16] geneplotter_1.64.0 knitr_1.29 Formula_1.2-3
[19] jsonlite_1.7.0 workflowr_1.6.2 broom_0.7.0
[22] annotate_1.64.0 cluster_2.1.0 dbplyr_1.4.4
[25] png_0.1-7 compiler_3.6.0 httr_1.4.1
[28] backports_1.1.8 assertthat_0.2.1 Matrix_1.2-18
[31] cli_2.0.2 later_1.1.0.1 acepack_1.4.1
[34] htmltools_0.5.0 tools_3.6.0 gtable_0.3.0
[37] glue_1.4.1 GenomeInfoDbData_1.2.2 Rcpp_1.0.5
[40] slam_0.1-47 cellranger_1.1.0 vctrs_0.3.1
[43] xfun_0.15 rvest_0.3.5 lifecycle_0.2.0
[46] XML_3.98-1.20 zlibbioc_1.32.0 scales_1.1.1
[49] hms_0.5.3 promises_1.1.1 RColorBrewer_1.1-2
[52] yaml_2.2.1 memoise_1.1.0 gridExtra_2.3
[55] rpart_4.1-15 latticeExtra_0.6-29 stringi_1.4.6
[58] RSQLite_2.2.0 genefilter_1.68.0 checkmate_2.0.0
[61] rlang_0.4.7 pkgconfig_2.0.3 bitops_1.0-6
[64] evaluate_0.14 lattice_0.20-41 lpsymphony_1.14.0
[67] labeling_0.3 htmlwidgets_1.5.1 bit_1.1-15.2
[70] tidyselect_1.1.0 magrittr_1.5 R6_2.4.1
[73] generics_0.0.2 Hmisc_4.4-0 DBI_1.1.0
[76] withr_2.2.0 pillar_1.4.6 haven_2.3.1
[79] foreign_0.8-71 survival_3.2-3 RCurl_1.98-1.2
[82] nnet_7.3-14 modelr_0.1.8 crayon_1.3.4
[85] fdrtool_1.2.15 rmarkdown_2.3 jpeg_0.1-8.1
[88] locfit_1.5-9.4 grid_3.6.0 readxl_1.3.1
[91] data.table_1.12.8 blob_1.2.1 git2r_0.27.1
[94] reprex_0.3.0 digest_0.6.25 xtable_1.8-4
[97] extraDistr_1.8.11 httpuv_1.5.4 munsell_0.5.0