Quality control of the new proteomic data from timsTOF machine
Junyan Lu
2020-02-27
Last updated: 2020-08-06
Checks: 6 1
Knit directory: Proteomics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200227) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3fb50c5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/analysisDrugResponses_IC50_cache/
Ignored: analysis/analysisDrugResponses_cache/
Ignored: analysis/complexAnalysis_IGHV_alternative_cache/
Ignored: analysis/complexAnalysis_IGHV_cache/
Ignored: analysis/complexAnalysis_trisomy12_alteredPQR_cache/
Ignored: analysis/complexAnalysis_trisomy12_alternative_cache/
Ignored: analysis/complexAnalysis_trisomy12_cache/
Ignored: analysis/correlateCLLPD_cache/
Ignored: analysis/correlateRNAexpression_cache/
Ignored: analysis/manuscript_S1_Overview_cache/
Ignored: analysis/manuscript_S2_genomicAssociation_cache/
Ignored: analysis/manuscript_S3_trisomy12_cache/
Ignored: analysis/manuscript_S4_IGHV_cache/
Ignored: analysis/manuscript_S5_trisomy19_cache/
Ignored: analysis/manuscript_S6_del11q_cache/
Ignored: analysis/manuscript_S7_SF3B1_cache/
Ignored: analysis/manuscript_S8_drugResponse_Outcomes_cache/
Ignored: analysis/predictOutcome_cache/
Ignored: code/.Rhistory
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/.trisomy12_norm.pdf
Untracked: analysis/CNVanalysis_11q.Rmd
Untracked: analysis/CNVanalysis_trisomy12.Rmd
Untracked: analysis/CNVanalysis_trisomy19.Rmd
Untracked: analysis/SUGP1_splicing.svg.xml
Untracked: analysis/analysisDrugResponses.Rmd
Untracked: analysis/analysisDrugResponses_IC50.Rmd
Untracked: analysis/analysisPCA.Rmd
Untracked: analysis/analysisSplicing.Rmd
Untracked: analysis/analysisTrisomy19.Rmd
Untracked: analysis/annotateCNV.Rmd
Untracked: analysis/complexAnalysis_IGHV.Rmd
Untracked: analysis/complexAnalysis_IGHV_alternative.Rmd
Untracked: analysis/complexAnalysis_overall.Rmd
Untracked: analysis/complexAnalysis_trisomy12.Rmd
Untracked: analysis/complexAnalysis_trisomy12_alternative.Rmd
Untracked: analysis/correlateGenomic_PC12adjusted.Rmd
Untracked: analysis/correlateGenomic_noBlock.Rmd
Untracked: analysis/correlateGenomic_noBlock_MCLL.Rmd
Untracked: analysis/correlateGenomic_noBlock_UCLL.Rmd
Untracked: analysis/correlateRNAexpression.Rmd
Untracked: analysis/default.css
Untracked: analysis/del11q.pdf
Untracked: analysis/del11q_norm.pdf
Untracked: analysis/manuscript_S0_PrepareData.Rmd
Untracked: analysis/manuscript_S1_Overview.Rmd
Untracked: analysis/manuscript_S2_genomicAssociation.Rmd
Untracked: analysis/manuscript_S3_trisomy12.Rmd
Untracked: analysis/manuscript_S4_IGHV.Rmd
Untracked: analysis/manuscript_S5_trisomy19.Rmd
Untracked: analysis/manuscript_S6_del11q.Rmd
Untracked: analysis/manuscript_S7_SF3B1.Rmd
Untracked: analysis/manuscript_S8_drugResponse_Outcomes.Rmd
Untracked: analysis/peptideValidate.Rmd
Untracked: analysis/plotExpressionCNV.Rmd
Untracked: analysis/processPeptides_LUMOS.Rmd
Untracked: analysis/processProteomics_timsTOF_Hela.Rmd
Untracked: analysis/processProteomics_timsTOF_new.Rmd
Untracked: analysis/qualityControl_timsTOF_new.Rmd
Untracked: analysis/style.css
Untracked: analysis/test.pdf
Untracked: analysis/test.svg
Untracked: analysis/trisomy12.pdf
Untracked: analysis/trisomy12_AFcor.Rmd
Untracked: analysis/trisomy12_norm.pdf
Untracked: code/AlteredPQR.R
Untracked: code/utils.R
Untracked: data/190909_CLL_prot_abund_med_norm.tsv
Untracked: data/190909_CLL_prot_abund_no_norm.tsv
Untracked: data/200725_cll_diaPASEF_direct_reports/
Untracked: data/200728_cll_diaPASEF_direct_plus_hela_reports/
Untracked: data/20190423_Proteom_submitted_samples_bereinigt.xlsx
Untracked: data/20191025_Proteom_submitted_samples_final.xlsx
Untracked: data/LUMOS/
Untracked: data/LUMOS_peptides/
Untracked: data/LUMOS_protAnnotation.csv
Untracked: data/LUMOS_protAnnotation_fix.csv
Untracked: data/SampleAnnotation_cleaned.xlsx
Untracked: data/example_proteomics_data
Untracked: data/facTab_IC50atLeast3New.RData
Untracked: data/gmts/
Untracked: data/mapEnsemble.txt
Untracked: data/mapSymbol.txt
Untracked: data/proteins_in_complexes
Untracked: data/pyprophet_export_aligned.csv
Untracked: data/timsTOF_protAnnotation.csv
Untracked: output/Fig1A.pdf
Untracked: output/Fig1A.png
Untracked: output/Fig1A.pptx
Untracked: output/LUMOS_processed.RData
Untracked: output/MSH6_splicing.svg
Untracked: output/SUGP1_splicing.eps
Untracked: output/SUGP1_splicing.pdf
Untracked: output/SUGP1_splicing.svg
Untracked: output/cnv_plots.zip
Untracked: output/cnv_plots/
Untracked: output/cnv_plots_norm.zip
Untracked: output/ddsrna_enc.RData
Untracked: output/deResList.RData
Untracked: output/deResList_timsTOF.RData
Untracked: output/dxdCLL.RData
Untracked: output/dxdCLL2.RData
Untracked: output/encMap.RData
Untracked: output/exprCNV.RData
Untracked: output/exprCNV_enc.RData
Untracked: output/lassoResults_CPS.RData
Untracked: output/lassoResults_IC50.RData
Untracked: output/patMeta_enc.RData
Untracked: output/pepCLL_lumos.RData
Untracked: output/pepCLL_lumos_enc.RData
Untracked: output/pepTab_lumos.RData
Untracked: output/pheno1000_enc.RData
Untracked: output/pheno1000_main.RData
Untracked: output/plotCNV_allChr11_diff.pdf
Untracked: output/plotCNV_del11q_sum.pdf
Untracked: output/proteomic_LUMOS_20200227.RData
Untracked: output/proteomic_LUMOS_20200320.RData
Untracked: output/proteomic_LUMOS_20200430.RData
Untracked: output/proteomic_LUMOS_enc.RData
Untracked: output/proteomic_timsTOF_20200227.RData
Untracked: output/proteomic_timsTOF_Hela_20200806.RData
Untracked: output/proteomic_timsTOF_enc.RData
Untracked: output/proteomic_timsTOF_new_20200806.RData
Untracked: output/splicingResults.RData
Untracked: output/survival_enc.RData
Untracked: output/timsTOF_processed.RData
Untracked: plotCNV_del11q_diff.pdf
Untracked: supp_latex/
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/analysisSF3B1.Rmd
Modified: analysis/compareProteomicsRNAseq.Rmd
Modified: analysis/correlateCLLPD.Rmd
Modified: analysis/correlateGenomic.Rmd
Deleted: analysis/correlateGenomic_removePC.Rmd
Modified: analysis/correlateMIR.Rmd
Modified: analysis/correlateMethylationCluster.Rmd
Modified: analysis/index.Rmd
Modified: analysis/predictOutcome.Rmd
Modified: analysis/processProteomics_LUMOS.Rmd
Modified: analysis/qualityControl_LUMOS.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Test association with RNA expression
Dimension of the inputed data
dim(protCLL)[1] 4977 50Process both datasets
colnames(dds) <- dds$PatID
dds <- estimateSizeFactors(dds)
sampleOverlap <- intersect(colnames(protCLL), colnames(dds))
geneOverlap <- intersect(rowData(protCLL)$ensembl_gene_id, rownames(dds))
ddsSub <- dds[geneOverlap, sampleOverlap]
protSub <- protCLL[match(geneOverlap, rowData(protCLL)$ensembl_gene_id), sampleOverlap]
#how many gene don't have RNA expression at all?
noExp <- rowSums(counts(ddsSub)) == 0
sum(noExp)[1] 9#remove those genes in both datasets
ddsSub <- ddsSub[!noExp,]
protSub <- protSub[!noExp,]
#remove proteins with duplicated identifiers
protSub <- protSub[!duplicated(rowData(protSub)$name)]
geneOverlap <- intersect(rowData(protSub)$ensembl_gene_id, rownames(ddsSub))
ddsSub.vst <- varianceStabilizingTransformation(ddsSub)Calculate correlations between protein abundance and RNA expression
rnaMat <- assay(ddsSub.vst)
proMat.raw <- assays(protSub)[["count"]]
proMat.qrilc <- assays(protSub)[["QRILC"]]
rownames(proMat.qrilc) <- rowData(protSub)$ensembl_gene_id
rownames(proMat.raw) <- rowData(protSub)$ensembl_gene_id
corTab <- lapply(geneOverlap, function(n) {
rna <- rnaMat[n,]
pro.q <- proMat.qrilc[n,]
pro.raw <- proMat.raw[n,]
res.q <- cor.test(rna, pro.q)
res.raw <- cor.test(rna, pro.raw, use = "pairwise.complete.obs")
tibble(id = n, impute=c("No Imputation","QRILC"),
p = c(res.raw$p.value, res.q$p.value),
coef = c(res.raw$estimate, res.q$estimate))
}) %>% bind_rows() %>%
arrange(desc(coef)) %>% mutate(p.adj = p.adjust(p, method = "BH"),
symbol = rowData(dds[id,])$symbol)Plot the distribution of correlation coefficient
ggplot(corTab, aes(x=coef, fill = impute)) + geom_histogram(position = "identity", col = "grey50", alpha =0.3, bins =100) +
geom_vline(xintercept = 0, col = "red", linetype = "dashed")Warning: Removed 2 rows containing non-finite values (stat_bin).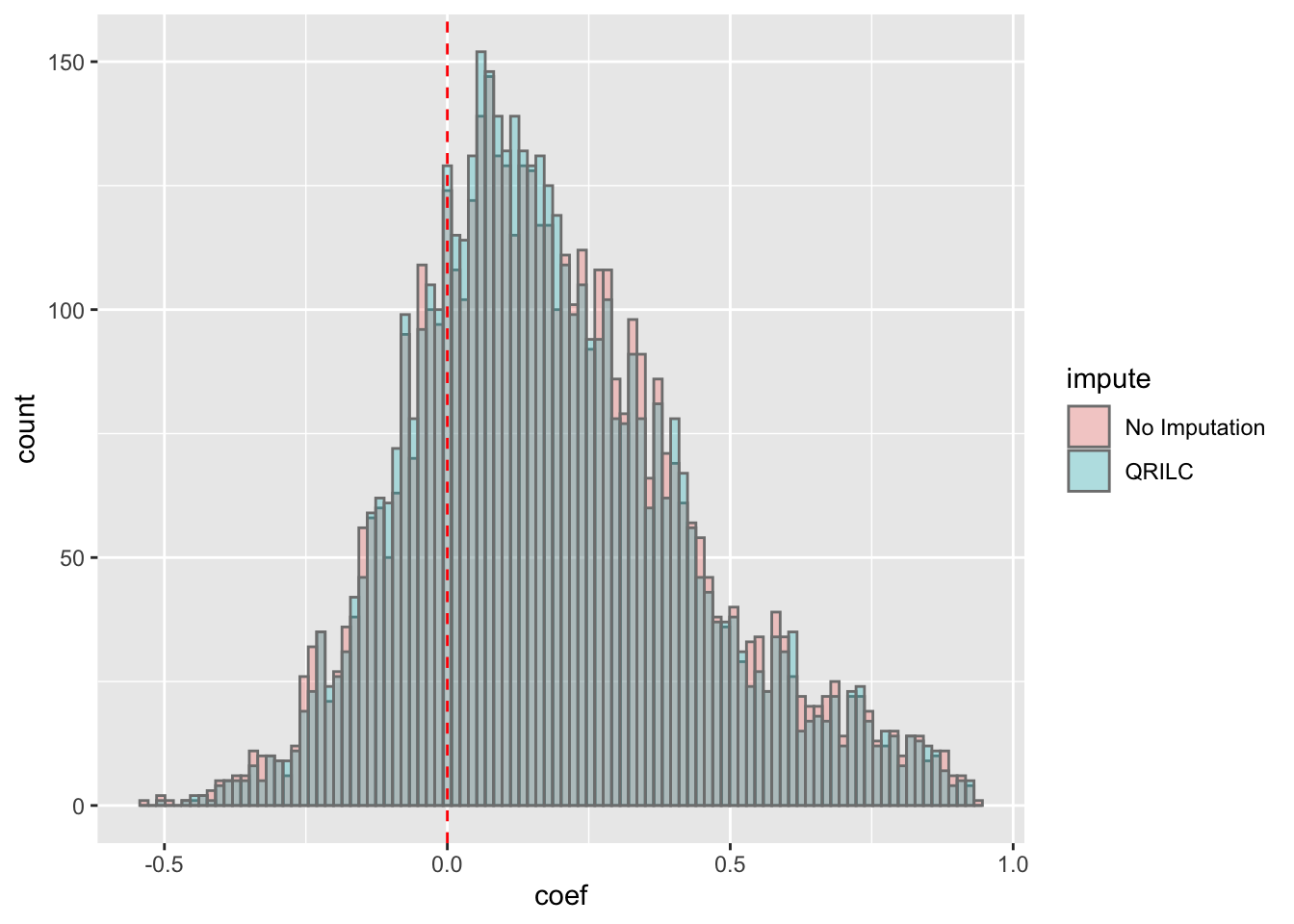 Most of the correlations are positive, which is reasonable.
Most of the correlations are positive, which is reasonable.
Number of significant positive and negative correlations (10% FDR)
sigTab <- corTab %>% filter(p.adj < 0.1) %>% mutate(direction = ifelse(coef > 0, "positive", "negative")) %>%
group_by(impute, direction) %>% summarise(number = length(id)) %>% ungroup() %>%
mutate(ratio = format(number/length(geneOverlap), digits = 2)) %>% arrange(number)`summarise()` regrouping output by 'impute' (override with `.groups` argument)DT::datatable(sigTab)Number of significant correlations VS FDR cut-off
plotTab <- lapply(seq(0,0.1, length.out = 100), function(fdr) {
filTab <- dplyr::filter(corTab, p.adj < fdr, coef > 0) %>%
group_by(impute) %>% summarise(n = length(id)) %>% mutate(fdr = fdr)
}) %>% bind_rows()`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)ggplot(plotTab, aes(x=fdr, y = n, col = impute))+ geom_line() +
ylab("Number of significant correlations") +
xlab("FDR cut-off")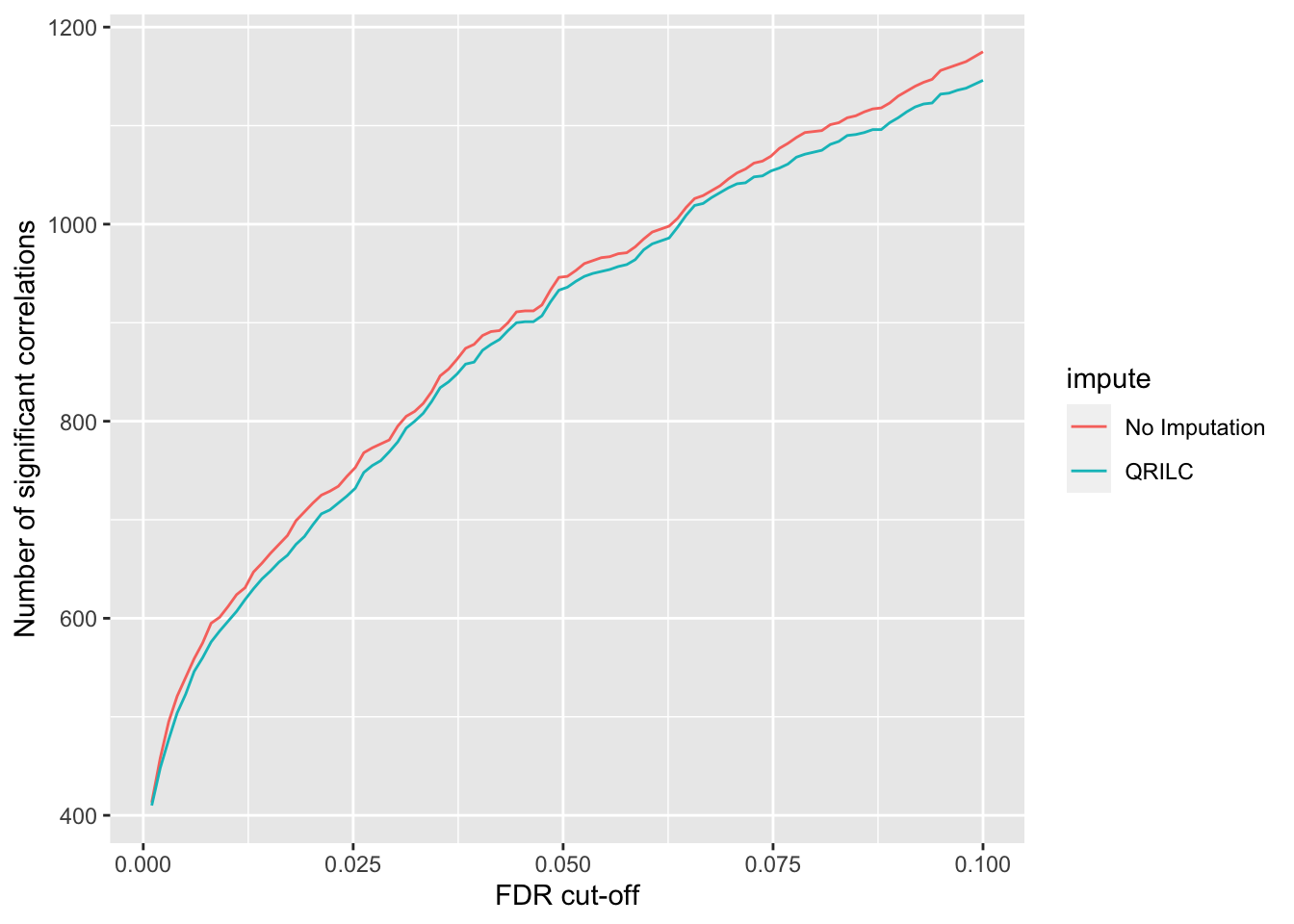
List of proteins that significantly correlated with RNA expression (10 %FDR, no imputation)
sigTab <- filter(corTab, p.adj < 0.1, impute == "No Imputation") %>% mutate_if(is.numeric, format, digits=2)
DT::datatable(sigTab)List of proteins that significantly correlated with RNA expression (10 %FDR, QRILC imputed)
sigTab <- filter(corTab, p.adj < 0.1, impute == "QRILC") %>% mutate_if(is.numeric, format, digits=2)
DT::datatable(sigTab)Correlation plot of top 9 most correlated protein-rna pairs
plotList <- lapply(sigTab$id[1:9], function(n) {
plotTab <- tibble(pro = proMat.qrilc[n,], gene = rnaMat[n,])
symbol <- filter(sigTab, id == n)$symbol
ggplot(plotTab, aes(x=pro, y=gene)) + geom_point() + geom_smooth(method = "lm") +
ggtitle(symbol) + ylab("RNA expression") + xlab("Protein abundance")
})
cowplot::plot_grid(plotlist = plotList, ncol =3)`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'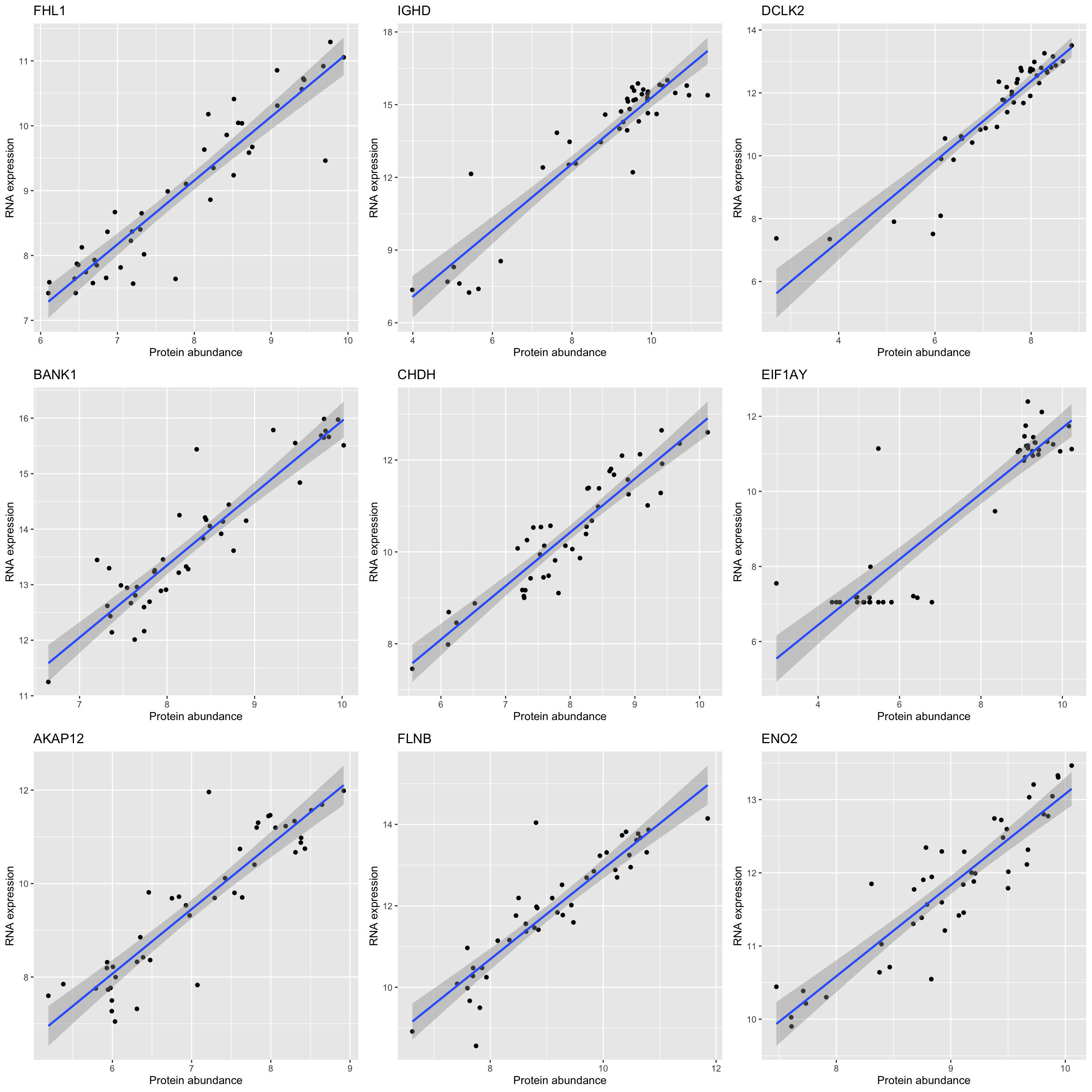
Assocation with technical factors
Are thechnical variables confounded with major genomic variabes?
# A tibble: 4 x 4
genomic technical pval p.adj
<chr> <chr> <dbl> <dbl>
1 IGHV.status processDate 0.0219 0.437
2 SF3B1 freeThawCycle 0.0229 0.437
3 del11q freeThawCycle 0.0369 0.437
4 trisomy12 processDate 0.0416 0.437No Significant assocations
Association between technical variables and priciple components of protein expression
plotMat <- assays(protCLL)[["QRILC"]]
pcRes <- prcomp(t(plotMat), center =TRUE, scale. = FALSE)$x
testRes <- lapply(colnames(pcRes), function(pc) {
lapply(colnames(techTab), function(tech) {
pcVar <- pcRes[,pc]
techVar <- techTab[[tech]]
res <- summary(aov(pcVar ~ techVar))
p <- res[[1]][1,4]
tibble(component = pc, technical = tech, pval = p)
}) %>% bind_rows()
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(pval, method = "BH")) %>%
arrange(pval)
filter(testRes, p.adj < 0.1)# A tibble: 3 x 4
component technical pval p.adj
<chr> <chr> <dbl> <dbl>
1 PC28 proteinConc 0.000102 0.0306
2 PC37 freeThawCycle 0.000298 0.0447
3 PC42 proteinConc 0.000575 0.0575There are some principle components correlated with technical variables. But the PCs are not top PCs, suggesting the know technical factor do not have impact on the major trends of the dataset.
Associations between technical variables and individual protein expressions
Association test
techTab <- colData(protCLL)[,c("operator", "viability","batch","processDate","proteinConc","freeThawCycle")] %>%
data.frame() %>%rownames_to_column("patID") %>% as_tibble() %>% mutate(processDate = as.character(processDate)) %>%
mutate_if(is.character, as.factor)%>% mutate_at(vars(-patID), as.numeric)
testTab <- assays(protCLL)[["QRILC"]] %>% data.frame() %>%
rownames_to_column("id") %>% mutate(name = rowData(protCLL)[id,]$hgnc_symbol) %>%
gather(key = "patID", value = "expr", -id, -name) %>%
left_join(techTab, by ="patID") %>% gather(key = technical, value = value, -id, -name, -patID, -expr)
testRes <- filter(testTab, !is.na(value)) %>%
group_by(name, technical) %>% nest() %>%
mutate(m = map(data, ~lm(expr~value,.))) %>%
mutate(res = map(m, broom::tidy)) %>%
unnest(res) %>% filter(term=="value") %>%
mutate(p.adj = p.adjust(p.value, method = "BH"))
sumTab <- filter(testRes, p.adj < 0.1) %>% group_by(technical) %>% summarise(n=length(name)) `summarise()` ungrouping output (override with `.groups` argument)ggplot(sumTab, aes(x=technical, y = n)) + geom_bar(stat = "identity") + coord_flip() +
xlab("") + ylab("Number of significantly associated proteins")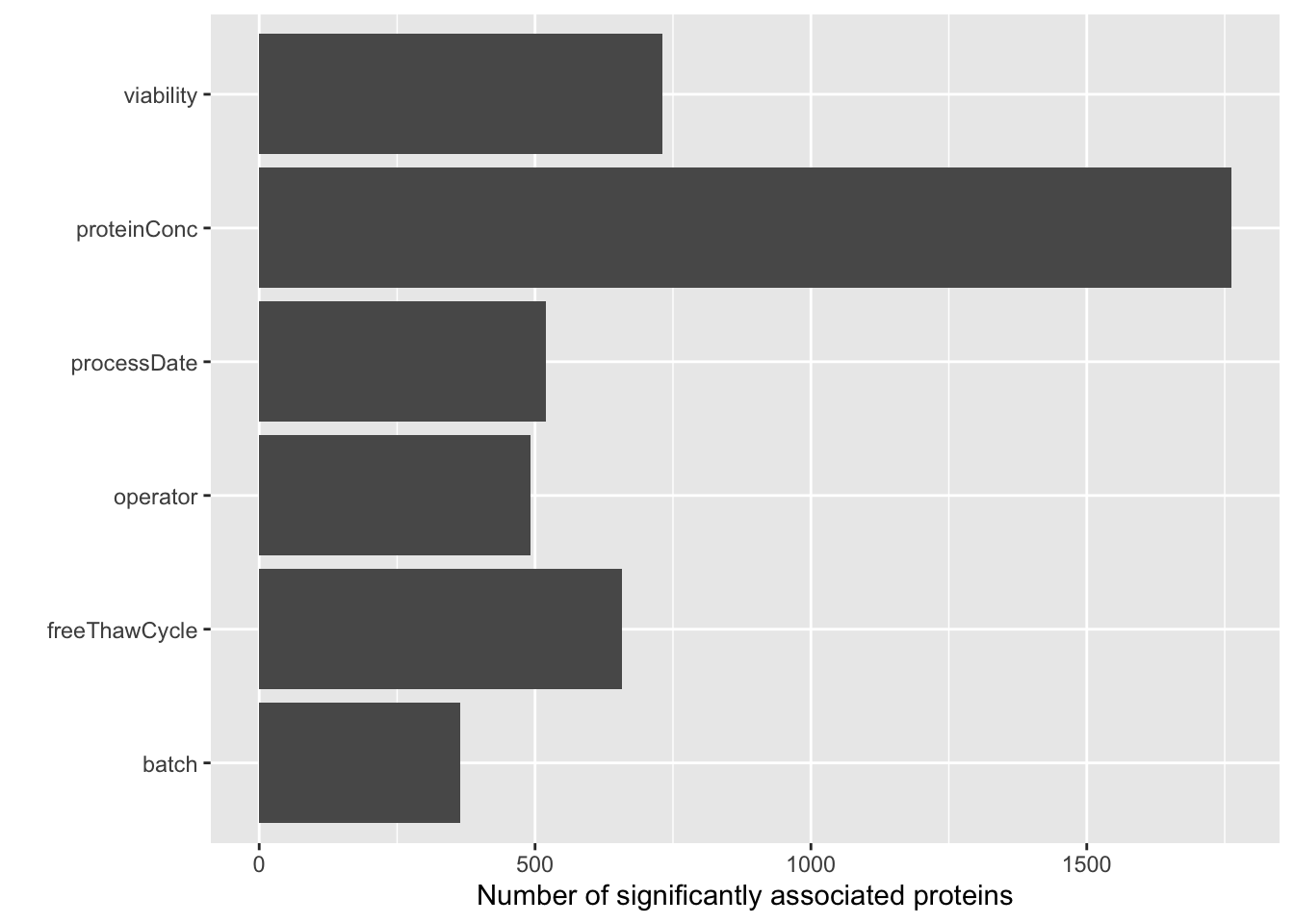
Assocation P value histogram for each technical factor
ggplot(testRes, aes(x=p.value)) + geom_histogram() + facet_wrap(~technical) +xlim(0,1)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 12 rows containing missing values (geom_bar).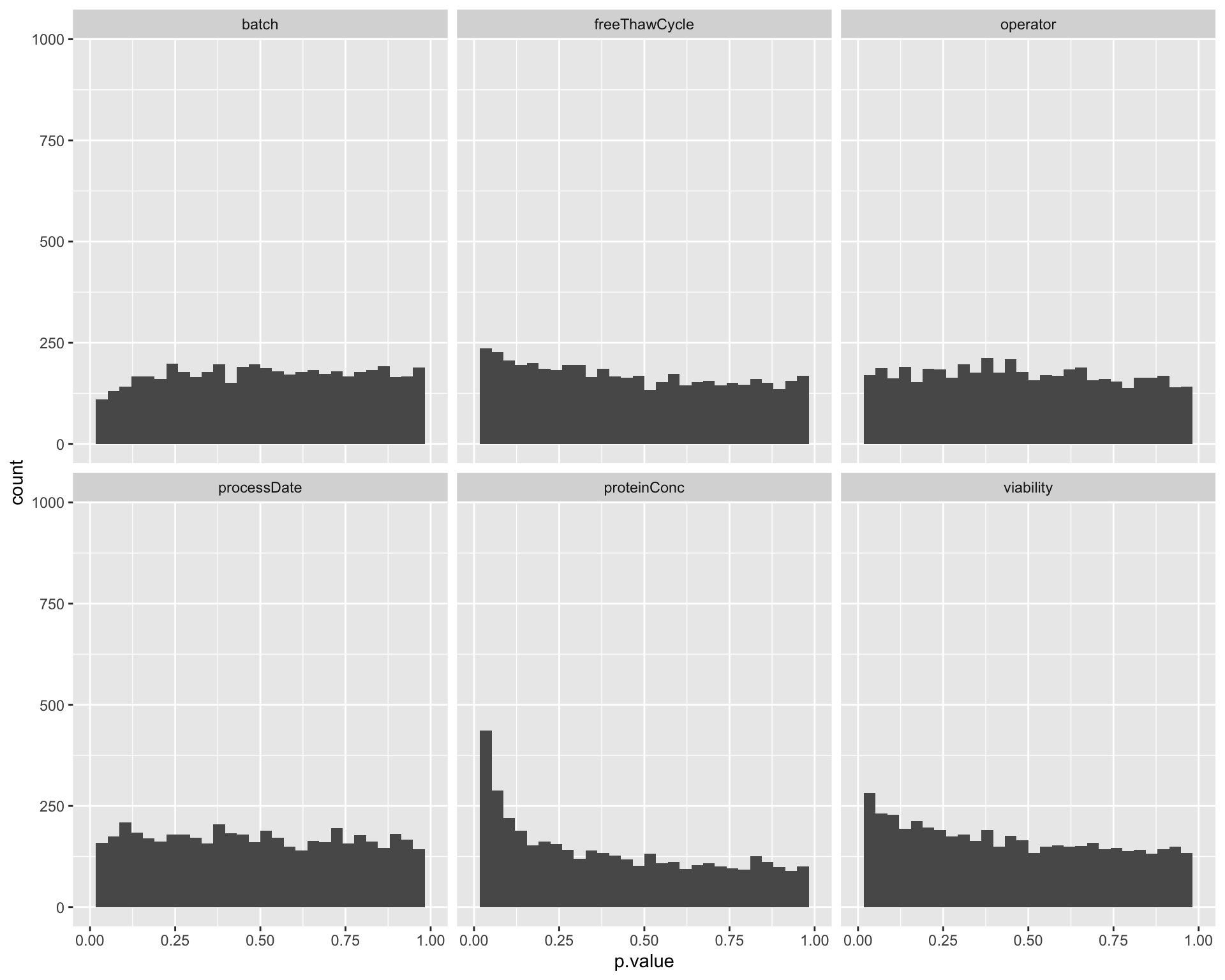 Also based on the p-value histogram, only overall protein concentration may have potential impact on protein abundance detection.
Also based on the p-value histogram, only overall protein concentration may have potential impact on protein abundance detection.
Will the correlation with RNA expression improve if we adjust for total protein concentration?
proteinConc <- techTab[match(colnames(proMat.qrilc), techTab$patID),]$proteinConc
corTab <- lapply(geneOverlap, function(n) {
rna <- rnaMat[n,]
pro.q <- proMat.qrilc[n,]
p.single <- anova(lm(rna ~ pro.q))$`Pr(>F)`[1]
p.multi <- car::Anova(lm(rna ~ pro.q + proteinConc))$`Pr(>F)`[1]
tibble(name = n, corrected = c("no","yes"),
p = c(p.single, p.multi))
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH")) %>% arrange(p)Number of significant correlations VS FDR cut-off
plotTab <- lapply(seq(0,0.1, length.out = 100), function(fdr) {
filTab <- dplyr::filter(corTab, p.adj < fdr) %>%
group_by(corrected) %>% summarise(n = length(name)) %>% mutate(fdr = fdr)
}) %>% bind_rows()`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)ggplot(plotTab, aes(x=fdr, y = n, col = corrected))+ geom_line() +
ylab("Number of significant correlations") +
xlab("FDR cut-off")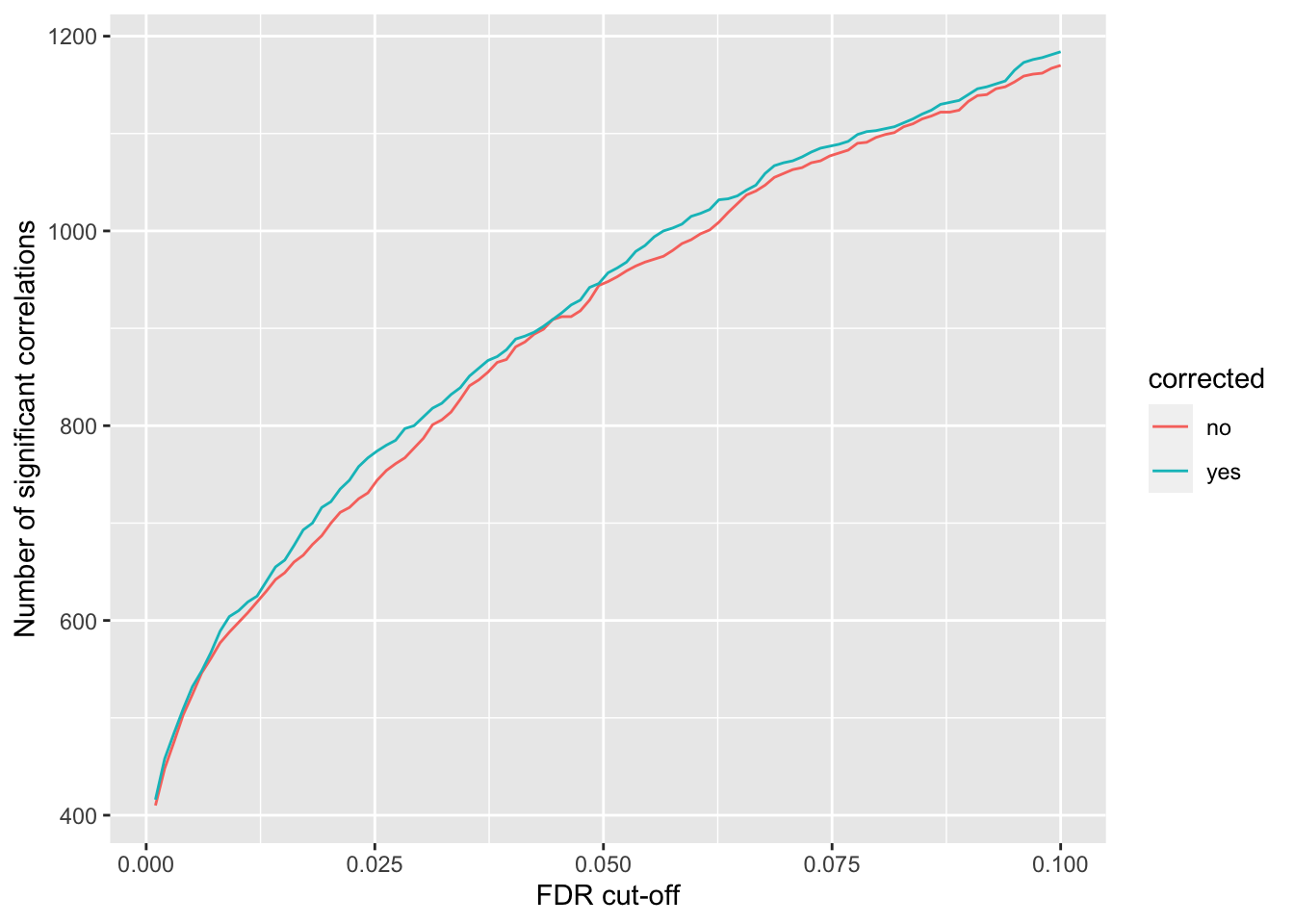 Seems to improve the correlation a little, but not much. We can include this factor in association test later.
Seems to improve the correlation a little, but not much. We can include this factor in association test later.
Check data structure
Hierarchical clustering
plotMat <- assays(protCLL)[["QRILC"]]
colAnno <- colData(protCLL)[,c("gender","IGHV.status","trisomy12")] %>%
data.frame()
plotMat <- jyluMisc::mscale(plotMat, censor = 6)
pheatmap(plotMat, scale = "none", annotation_col = colAnno, clustering_method = "ward.D2",
show_rownames = FALSE, color = colorRampPalette(c("navy","white","firebrick"))(100),
breaks = seq(-6,6, length.out = 101))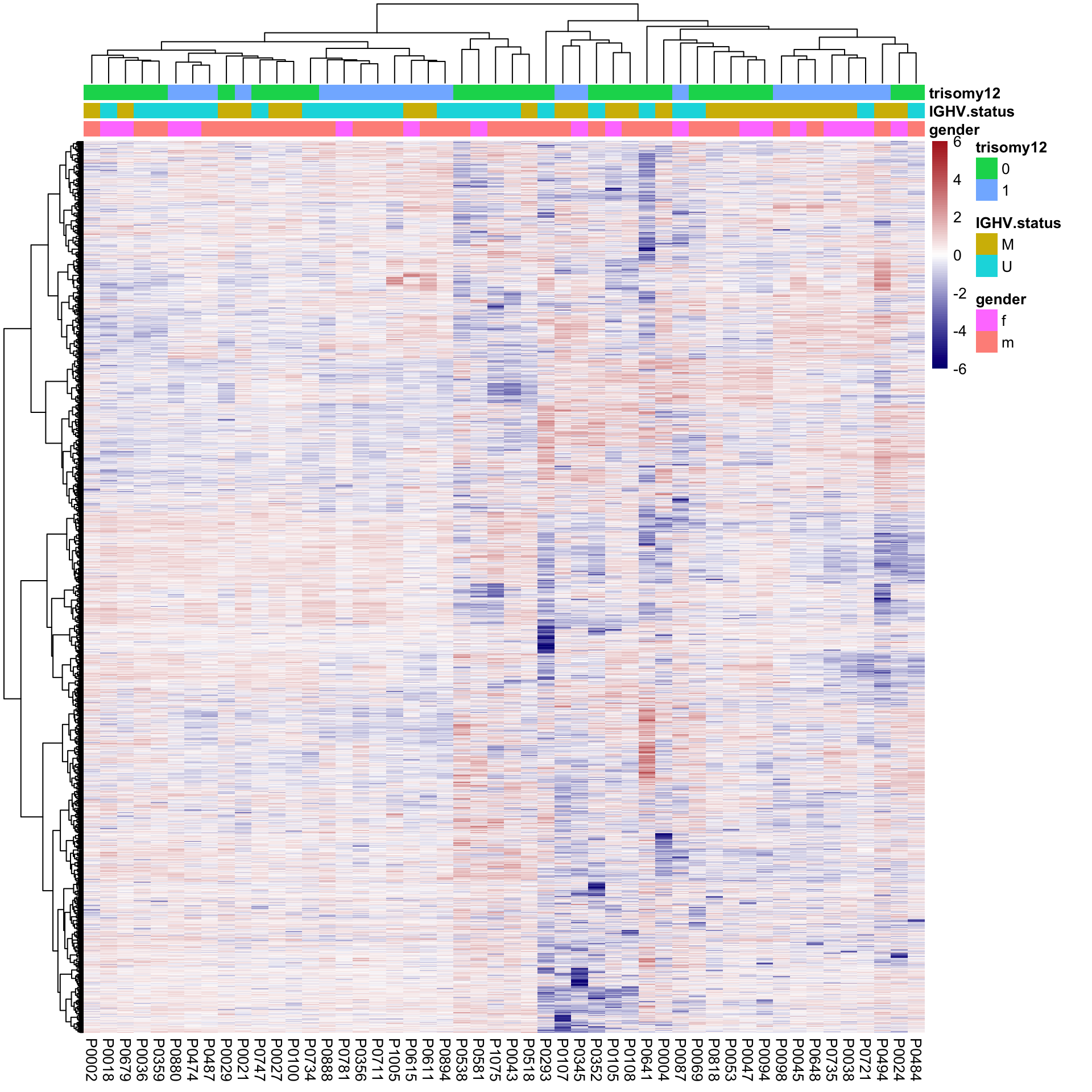 No clear separation can be observed
No clear separation can be observed
PCA
pcOut <- prcomp(t(plotMat), center =TRUE, scale. = FALSE)
pcRes <- pcOut$x
eigs <- pcOut$sdev^2
varExp <- structure(eigs/sum(eigs),names = colnames(pcRes))
plotTab <- pcRes[,1:2] %>% data.frame() %>% cbind(colAnno[rownames(.),]) %>%
rownames_to_column("patID") %>% as_tibble()
ggplot(plotTab, aes(x=PC1, y=PC2, col = IGHV.status, shape = trisomy12)) + geom_point(size=4) +
xlab(sprintf("PC1 (%1.2f%%)",varExp[["PC1"]]*100)) +
ylab(sprintf("PC2 (%1.2f%%)",varExp[["PC2"]]*100))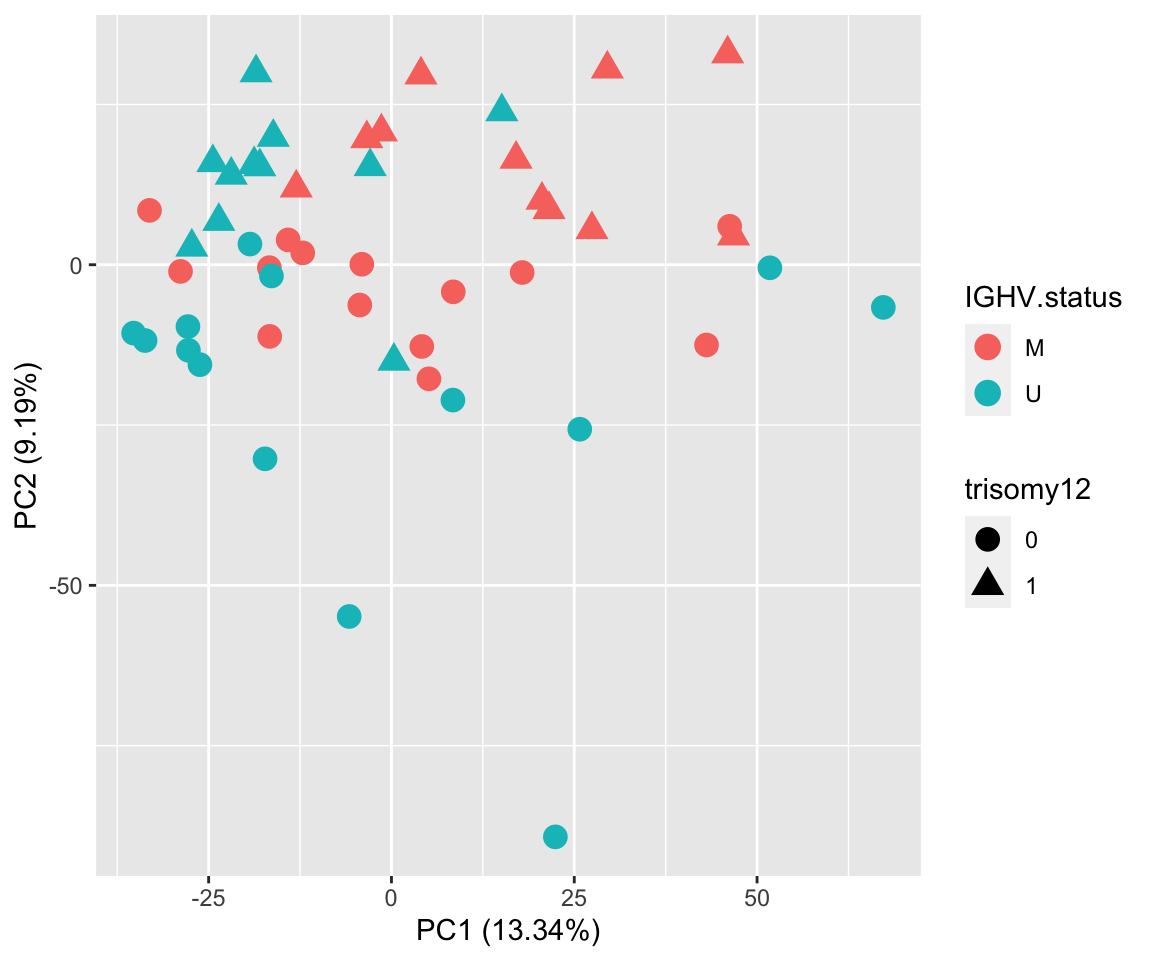 PC2 separates trisomy12
PC2 separates trisomy12
Correlation PCs with trisomy12 and IGHV status
corTab <- lapply(colnames(pcRes), function(pc) {
ighvCor <- t.test(pcRes[,pc] ~ colAnno$IGHV.status)
tri12Cor <- t.test(pcRes[,pc] ~ colAnno$trisomy12)
tibble(PC = pc,
feature=c("IGHV", "trisomy12"),
p = c(ighvCor$p.value, tri12Cor$p.value))
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH")) %>%
filter(p <= 0.05) %>% arrange(p)
corTab# A tibble: 7 x 4
PC feature p p.adj
<chr> <chr> <dbl> <dbl>
1 PC2 trisomy12 0.000000234 0.0000234
2 PC5 IGHV 0.000142 0.00708
3 PC6 trisomy12 0.00207 0.0691
4 PC4 trisomy12 0.00420 0.105
5 PC7 IGHV 0.0201 0.402
6 PC1 IGHV 0.0355 0.566
7 PC5 trisomy12 0.0396 0.566 PC6 is for IGHV
PCA plot using PC2 and PC7
plotTab <- pcRes[,1:10] %>% data.frame() %>% cbind(colAnno[rownames(.),]) %>%
rownames_to_column("patID") %>% as_tibble()
ggplot(plotTab, aes(x=PC2, y=PC6, col = IGHV.status, shape = trisomy12, label = patID)) + geom_point() + ggrepel::geom_text_repel() +
xlab(sprintf("PC2 (%1.2f%%)",varExp[["PC2"]]*100)) +
ylab(sprintf("PC6 (%1.2f%%)",varExp[["PC6"]]*100))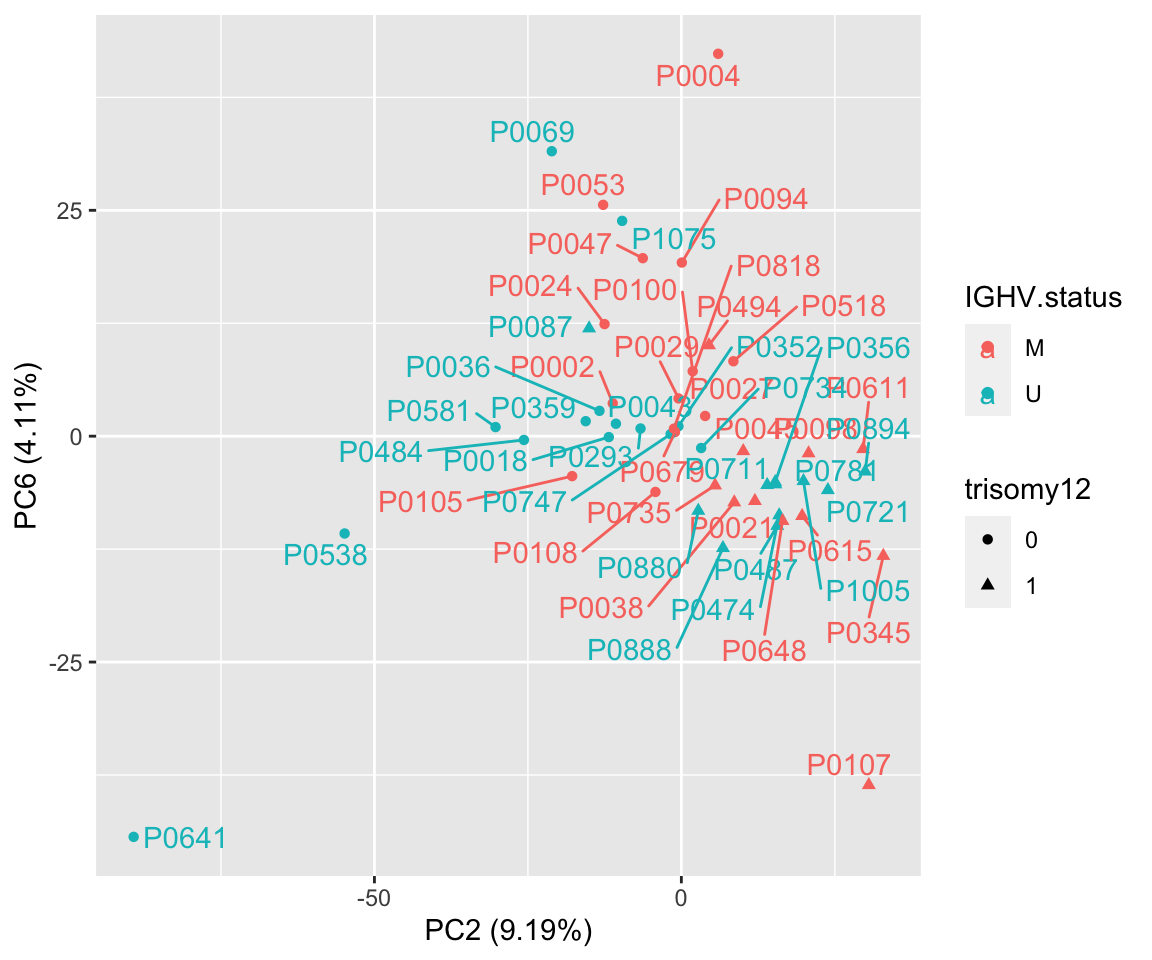 Using PC3 and PC4 better seperates IGHV and trisomy12.
Using PC3 and PC4 better seperates IGHV and trisomy12.
Biological annotation of PC1
Annotate PC1
Correlation test
Assocation test
proMat <- assays(protCLL)[["QRILC"]]
pc <- pcRes[,1][colnames(proMat)]
designMat <- model.matrix(~1+pc)
fit <- lmFit(proMat, designMat)
fit2 <- eBayes(fit)
corRes.pc1 <- topTable(fit2, number ="all", adjust.method = "BH", coef = "pc") %>% rownames_to_column("id") %>%
mutate(symbol = rowData(protCLL[id,])$hgnc_symbol)Number of significant associations (10% FDR)
hist(corRes.pc1$P.Value,breaks=100)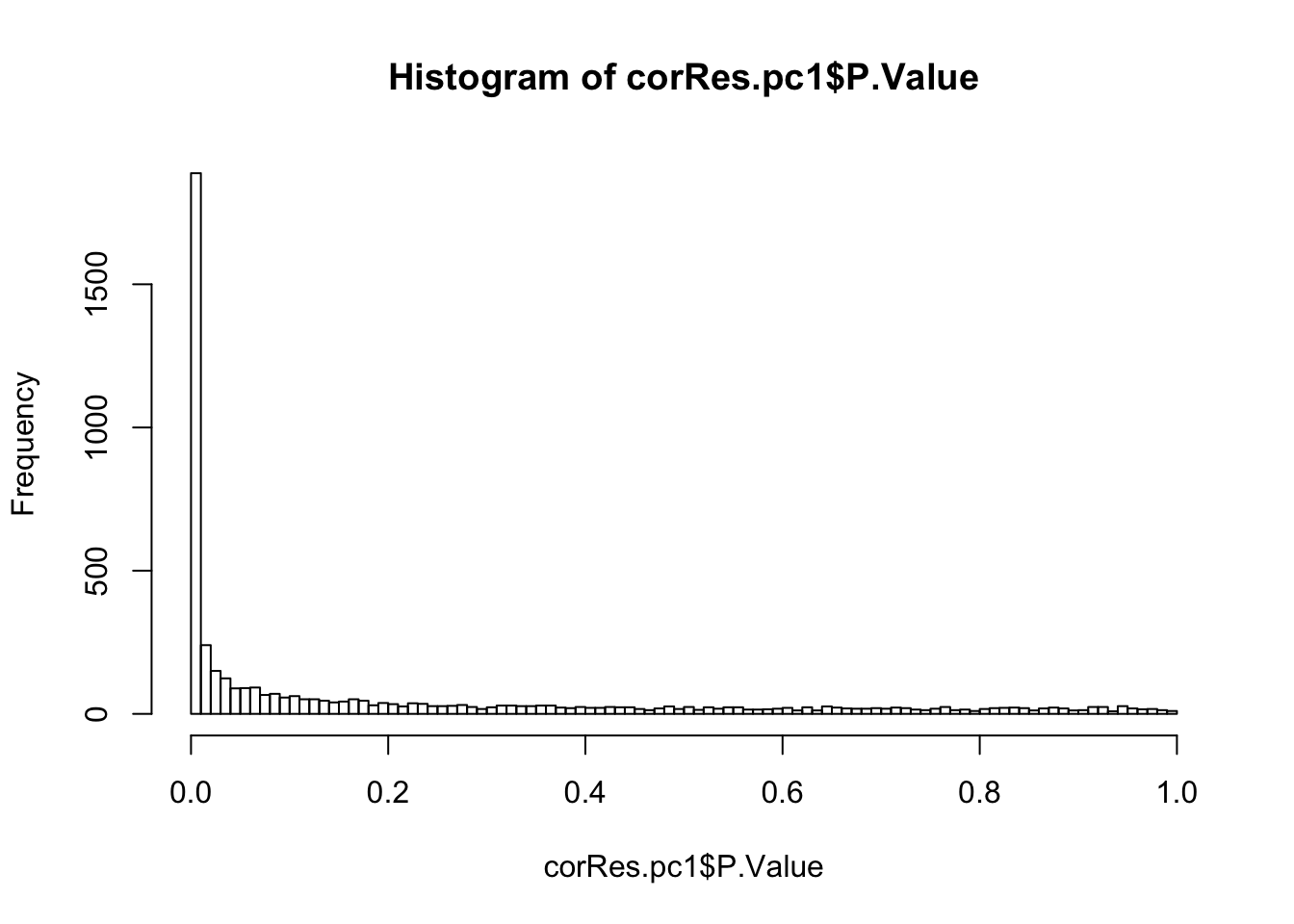
Table of significant associations (5% FDR)
resTab.sig <- filter(corRes.pc1, adj.P.Val < 0.05) %>%
select(symbol, id,logFC, P.Value, adj.P.Val) %>%
arrange(P.Value)
resTab.sig %>% mutate_if(is.numeric, formatC, digits=2, format= "e") %>%
DT::datatable()Heatmap of associated genes
colAnno <- tibble(patID = colnames(proMat), PC1 = pcRes[colnames(proMat),1]) %>%
arrange(PC1) %>% data.frame() %>% column_to_rownames("patID")
plotMat <- proMat[resTab.sig$id[1:100],rownames(colAnno)]
plotMat <- jyluMisc::mscale(plotMat, censor = 6)
pheatmap(plotMat, scale = "none", annotation_col = colAnno, clustering_method = "ward.D2",
cluster_cols = FALSE,
labels_row = resTab.sig$symbol[1:100], color = colorRampPalette(c("navy","white","firebrick"))(100),
breaks = seq(-6,6, length.out = 101))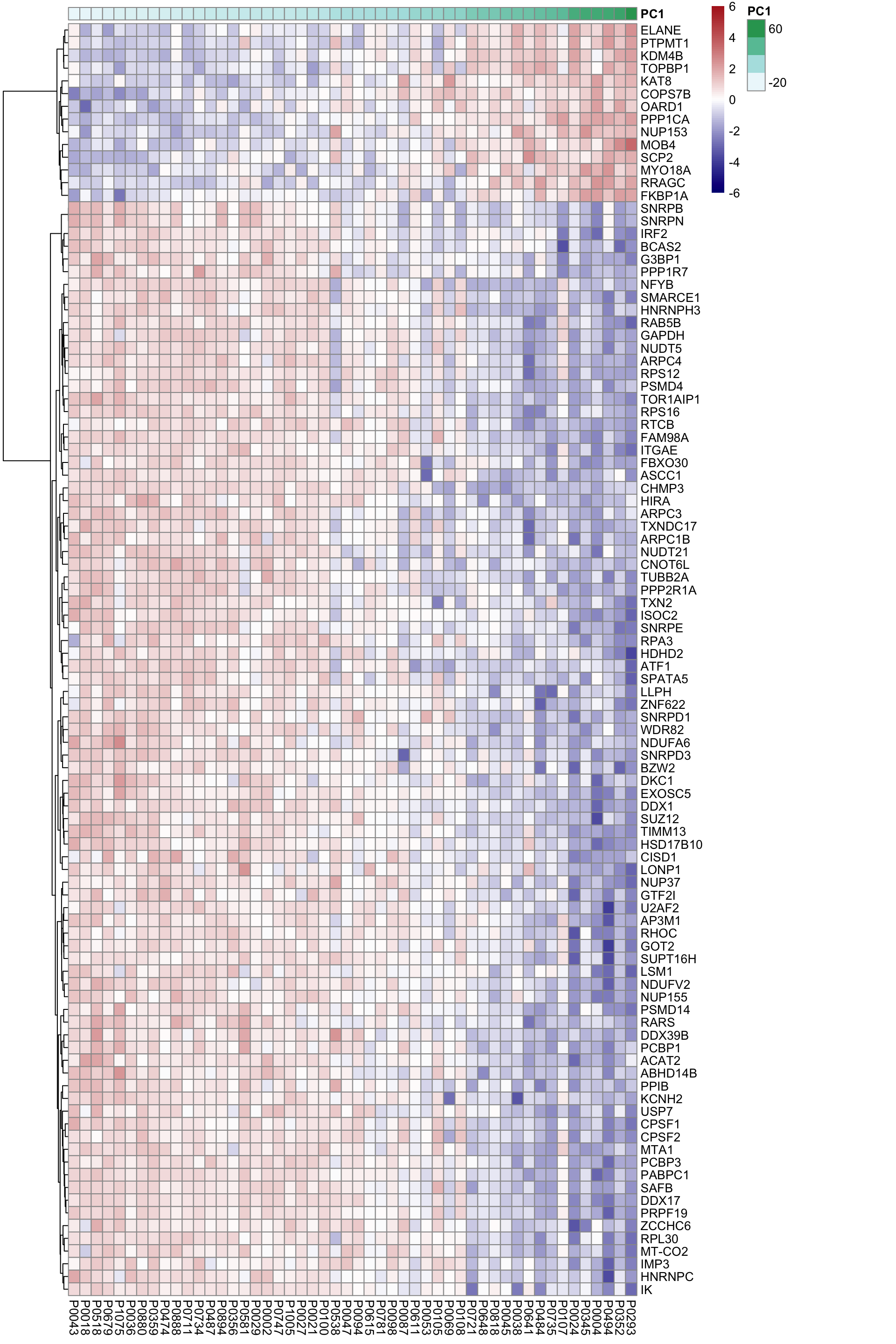
Enrichment using Camera
Hallmarks
gmts = list(H= "../data/gmts/h.all.v6.2.symbols.gmt",
C6 = "../data/gmts/c6.all.v6.2.symbols.gmt",
KEGG = "../data/gmts/c2.cp.kegg.v6.2.symbols.gmt")
res <- runCamera(proMat, designMat, gmts$H, id = rowData(protCLL[rownames(proMat),])$hgnc_symbol)
res$enrichPlot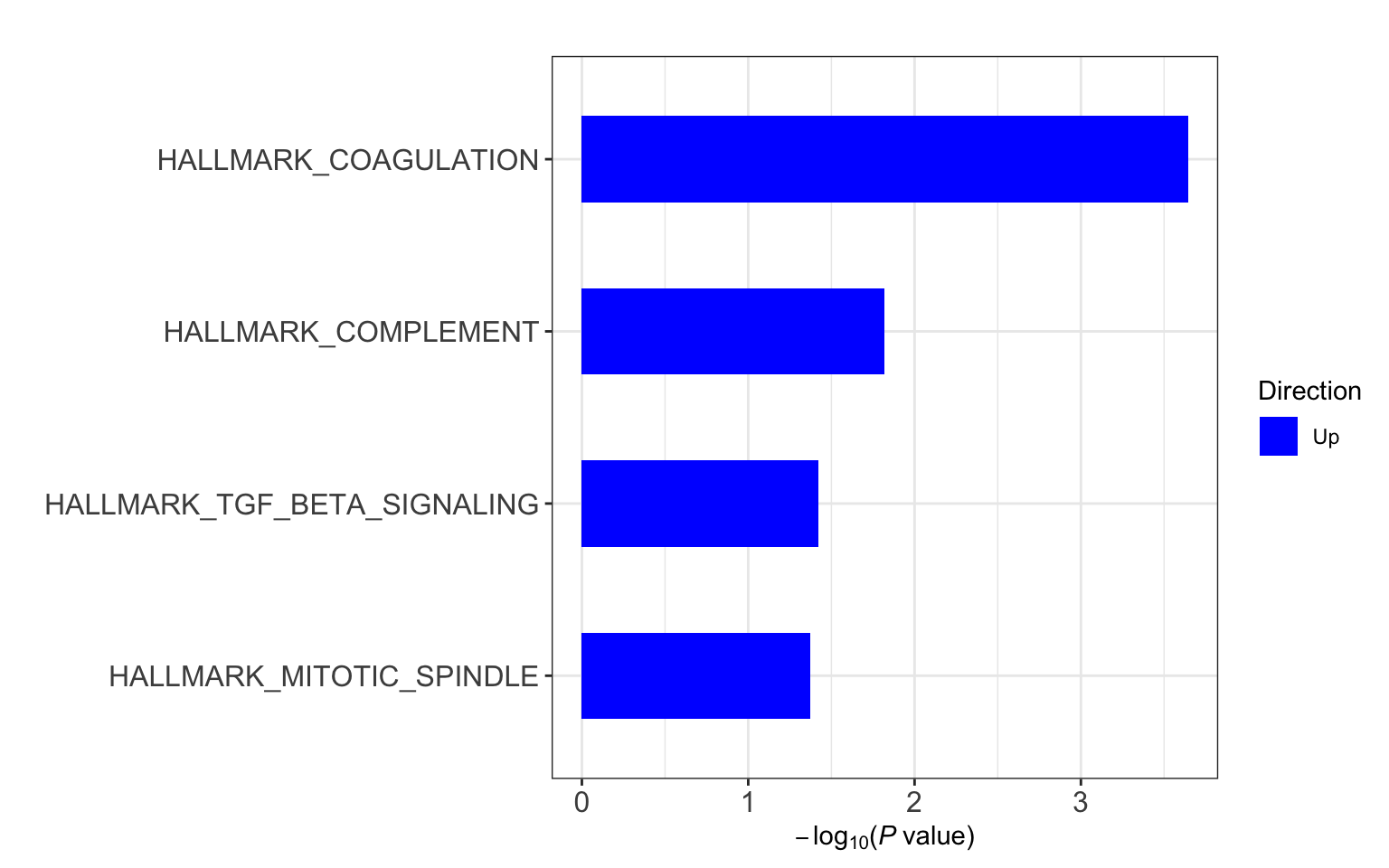
C6
res <- runCamera(proMat, designMat, gmts$C6, id = rowData(protCLL[rownames(proMat),])$hgnc_symbol)
res$enrichPlot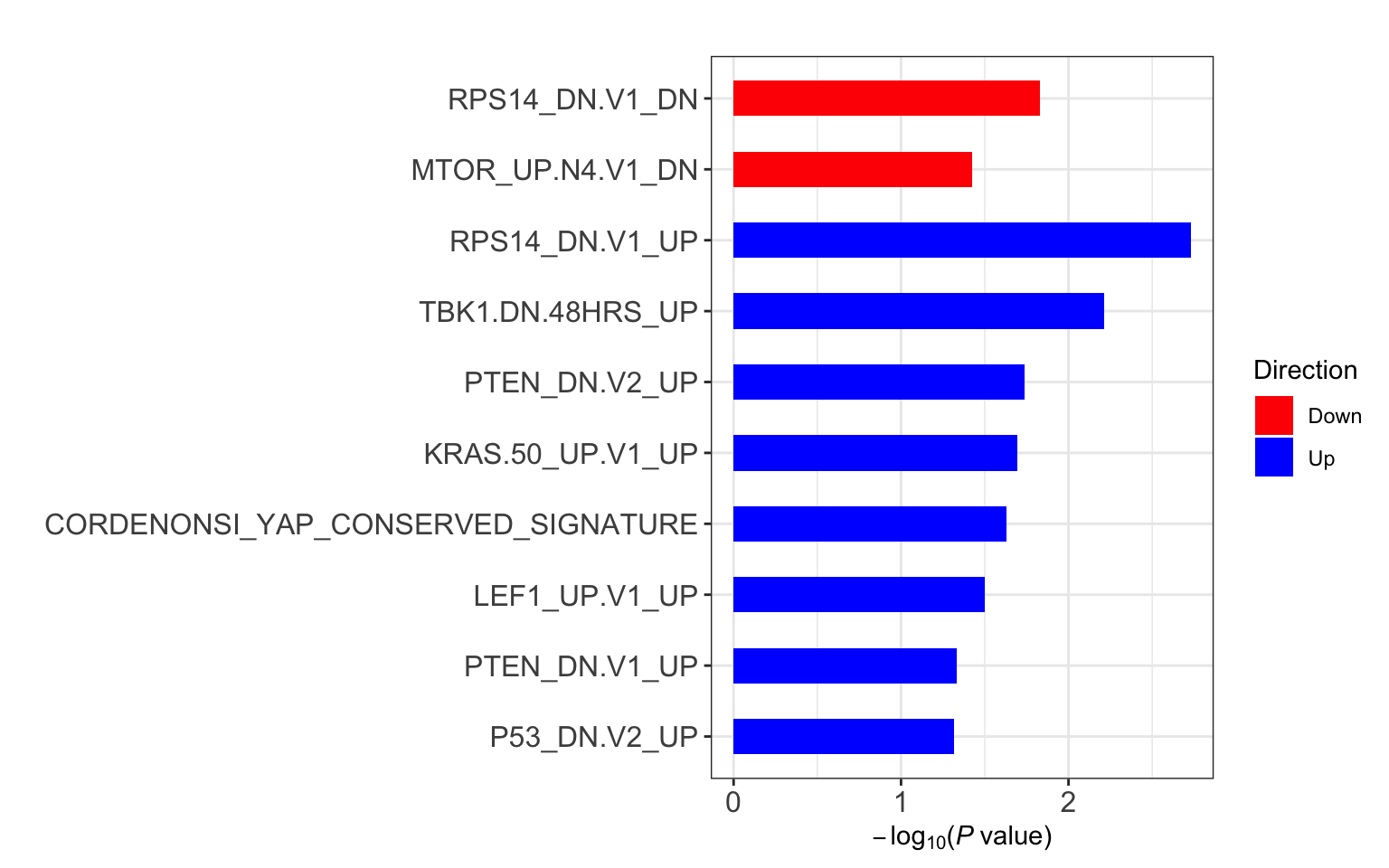
KEGG
res <- runCamera(proMat, designMat, gmts$KEGG, id = rowData(protCLL[rownames(proMat),])$hgnc_symbol)
res$enrichPlot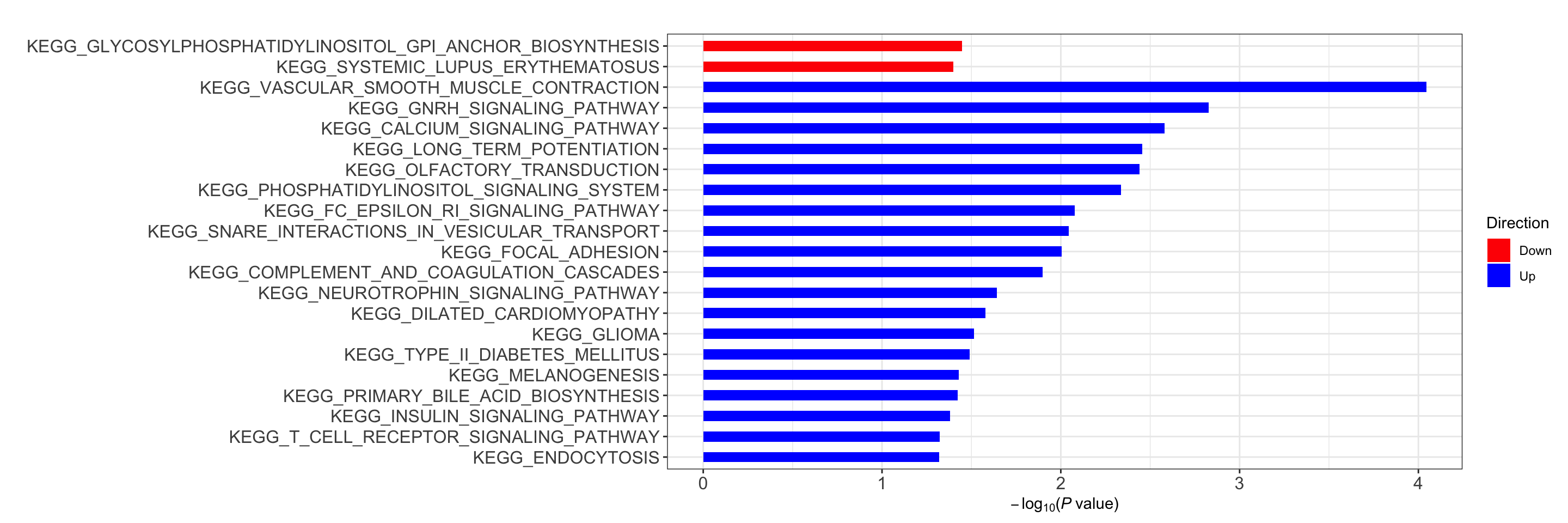
Association with lymphocyte count (potential contamination?)
load("~/CLLproject_jlu/ShinyApps/sampleTimeline//timeline.RData")
plotTab <- pcRes[,1:9] %>% data.frame() %>%
rownames_to_column("patID") %>% as_tibble() %>%
mutate(sampleID = protCLL[,patID]$sampleID) %>%
mutate(lymCount = sampleTab[match(sampleID, sampleTab$sampleID),]$leukCount) %>%
gather(key = "pc", value = "val",-patID,-sampleID,-lymCount)
ggplot(plotTab, aes(x=val, y=lymCount)) + geom_point() + geom_smooth(method = "lm") +
facet_wrap(~pc, ncol =3, scale = "free")`geom_smooth()` using formula 'y ~ x'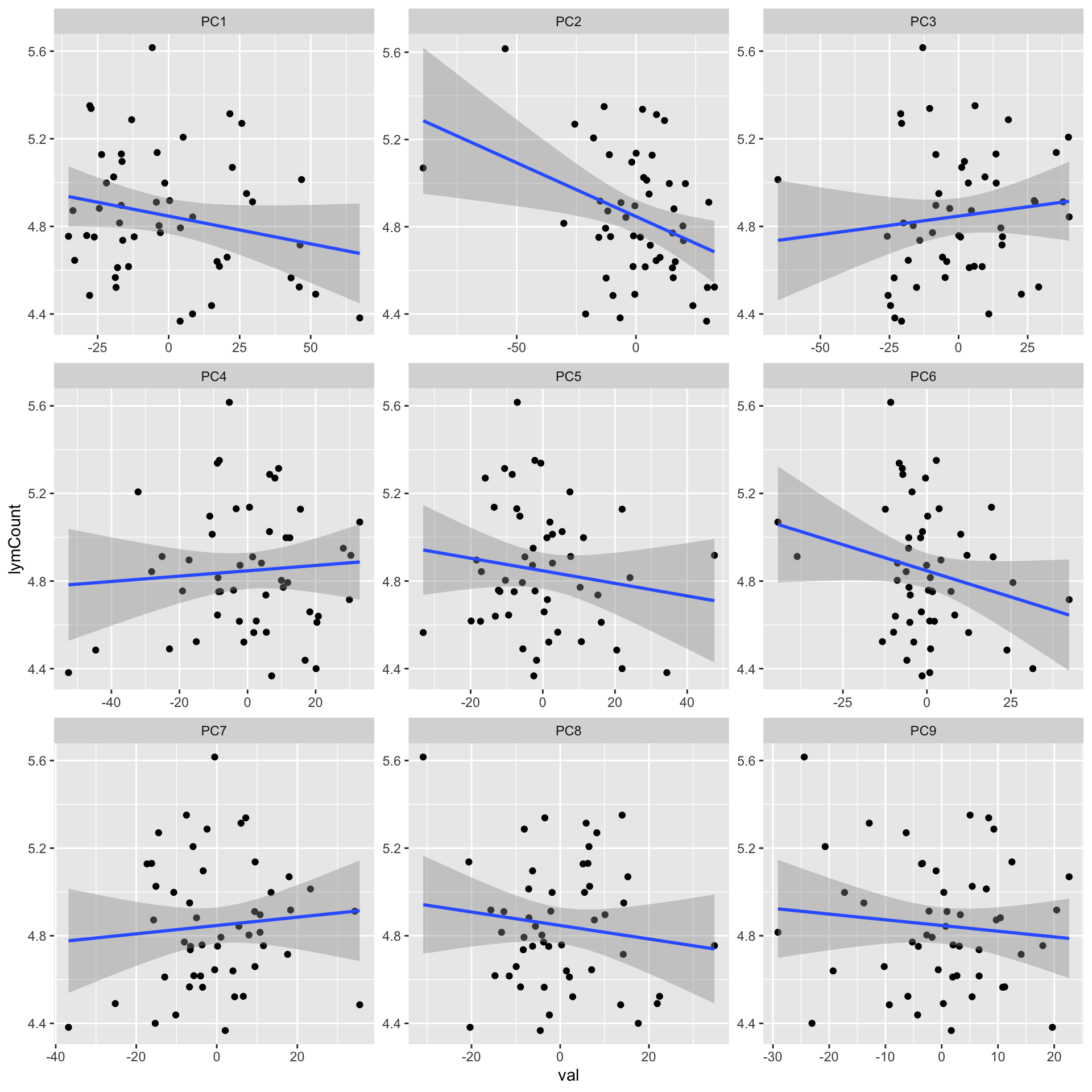
corRes <- plotTab %>% group_by(pc) %>% nest() %>%
mutate(m= map(data, ~cor.test(~ val+ lymCount,.))) %>%
mutate(res = map(m, broom::tidy)) %>%
unnest(res) %>% select(pc, estimate, p.value) %>%
arrange(p.value)
corRes# A tibble: 9 x 3
# Groups: pc [9]
pc estimate p.value
<chr> <dbl> <dbl>
1 PC2 -0.364 0.00927
2 PC6 -0.236 0.0987
3 PC1 -0.226 0.115
4 PC5 -0.145 0.315
5 PC8 -0.131 0.364
6 PC3 0.122 0.399
7 PC9 -0.105 0.467
8 PC7 0.0908 0.530
9 PC4 0.0766 0.597 Association wit %Lymphocyte PB
library(DBI)
con <- dbConnect(RPostgreSQL::PostgreSQL(),
dbname = "tumorbank",
host = "huber-vm01.embl.de",
user = "admin",
password = "bloodcancertumorbank")
PBtab <- tbl(con, "patient") %>%
left_join(tbl(con, "sample"), by = c(patid = "smppatidref")) %>%
left_join(tbl(con, "analysis"), by = c(smpid = "anlsmpidref")) %>%
collect() %>%
select(patpatientid, smpsampleid, smpleukocytes, smpsampledate, smppblymphocytes)
dbDisconnect(con)[1] TRUEplotTab <- pcRes[,1:9] %>% data.frame() %>%
rownames_to_column("patID") %>% as_tibble() %>%
mutate(sampleID = protCLL[,patID]$sampleID) %>%
mutate(perPB = PBtab[match(sampleID, PBtab$smpsampleid),]$smppblymphocytes) %>%
gather(key = "pc", value = "val",-patID,-sampleID,-perPB)
ggplot(plotTab, aes(x=val, y=perPB)) + geom_point() + geom_smooth(method = "lm") +
facet_wrap(~pc, ncol =3, scale = "free")`geom_smooth()` using formula 'y ~ x'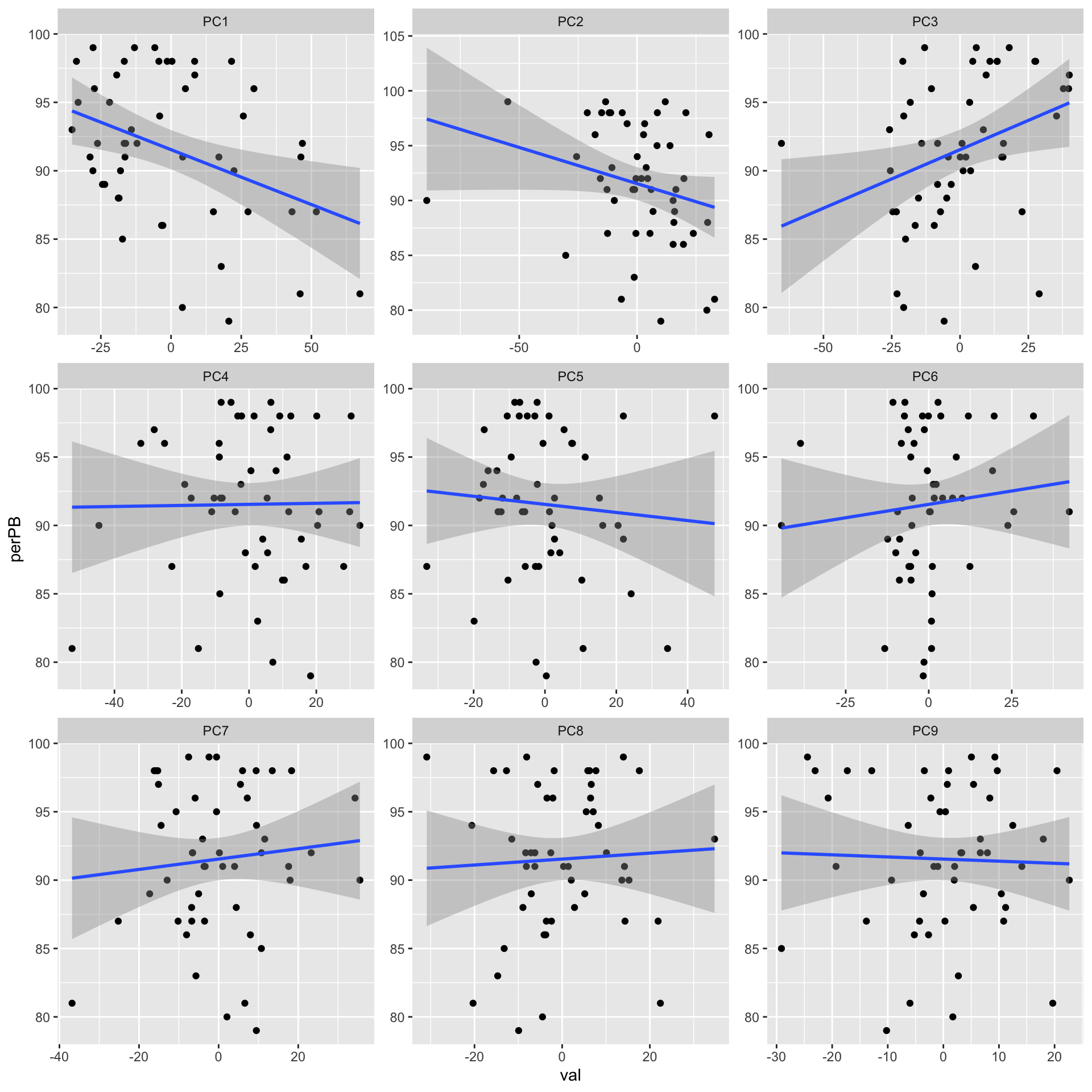
corRes <- plotTab %>% group_by(pc) %>% nest() %>%
mutate(m= map(data, ~cor.test(~ val+ perPB,.))) %>%
mutate(res = map(m, broom::tidy)) %>%
unnest(res) %>% select(pc, estimate, p.value) %>%
arrange(p.value)
corRes# A tibble: 9 x 3
# Groups: pc [9]
pc estimate p.value
<chr> <dbl> <dbl>
1 PC1 -0.382 0.00626
2 PC3 0.328 0.0199
3 PC2 -0.260 0.0679
4 PC6 0.103 0.475
5 PC7 0.0967 0.504
6 PC5 -0.0799 0.581
7 PC8 0.0494 0.733
8 PC9 -0.0334 0.818
9 PC4 0.0131 0.928 PC1 and PC2 both seem to correlation with %PB. But PC2 is also associate with trisomy12?
Will the correlation with transcriptomics increase if PC1 is regressed out?
pc1 <- pcRes[colnames(rnaMat),1]
corTab <- lapply(geneOverlap, function(n) {
rna <- rnaMat[n,]
pro.q <- proMat.qrilc[n,]
p.single <- anova(lm(rna ~ pro.q))$`Pr(>F)`[1]
p.multi <- car::Anova(lm(rna ~ pro.q + pc1))$`Pr(>F)`[1]
tibble(name = n, corrected = c("no","yes"),
p = c(p.single, p.multi))
}) %>% bind_rows() %>% mutate(p.adj = p.adjust(p, method = "BH")) %>% arrange(p)Number of significant correlations VS FDR cut-off
plotTab <- lapply(seq(0,0.1, length.out = 100), function(fdr) {
filTab <- dplyr::filter(corTab, p.adj < fdr) %>%
group_by(corrected) %>% summarise(n = length(name)) %>% mutate(fdr = fdr)
}) %>% bind_rows()`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)
`summarise()` ungrouping output (override with `.groups` argument)ggplot(plotTab, aes(x=fdr, y = n, col = corrected))+ geom_line() +
ylab("Number of significant correlations") +
xlab("FDR cut-off")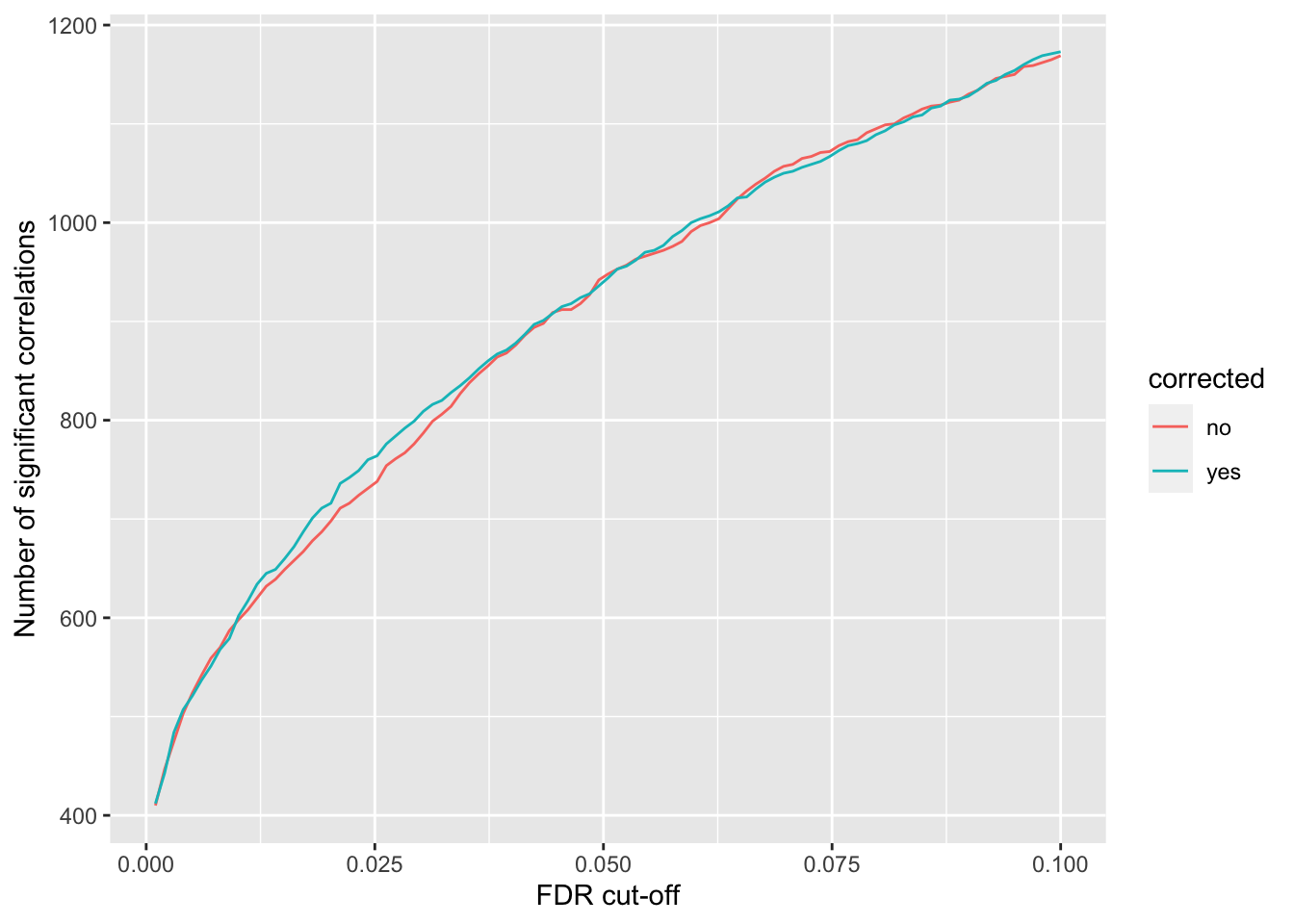 Seems to improve the correlation a little, but not much. We can include this factor in association test later.
Seems to improve the correlation a little, but not much. We can include this factor in association test later.
Reconstruct data with PC1 and PC2 removed
protMat <- t(assays(protCLL)[["QRILC"]])
mu <- colMeans(protMat)
Xpca <- prcomp(protMat, center = TRUE, scale. = FALSE)
#reconstruct without the first two components
protMat.new <- Xpca$x[,2:ncol(Xpca$x)] %*% t(Xpca$rotation[,2:ncol(Xpca$x)])
protMat.new <- scale(protMat.new, center = -mu, scale = FALSE)
protMat.new <- t(protMat.new)Hierarchical clustering using reconstructed matrix
plotMat <- protMat.new
colAnno <- colData(protCLL)[,c("gender","IGHV.status","trisomy12")] %>%
data.frame()
plotMat <- jyluMisc::mscale(plotMat, censor = 6)
pheatmap(plotMat, scale = "none", annotation_col = colAnno, clustering_method = "ward.D2",
show_rownames = FALSE, color = colorRampPalette(c("navy","white","firebrick"))(100),
breaks = seq(-6,6, length.out = 101))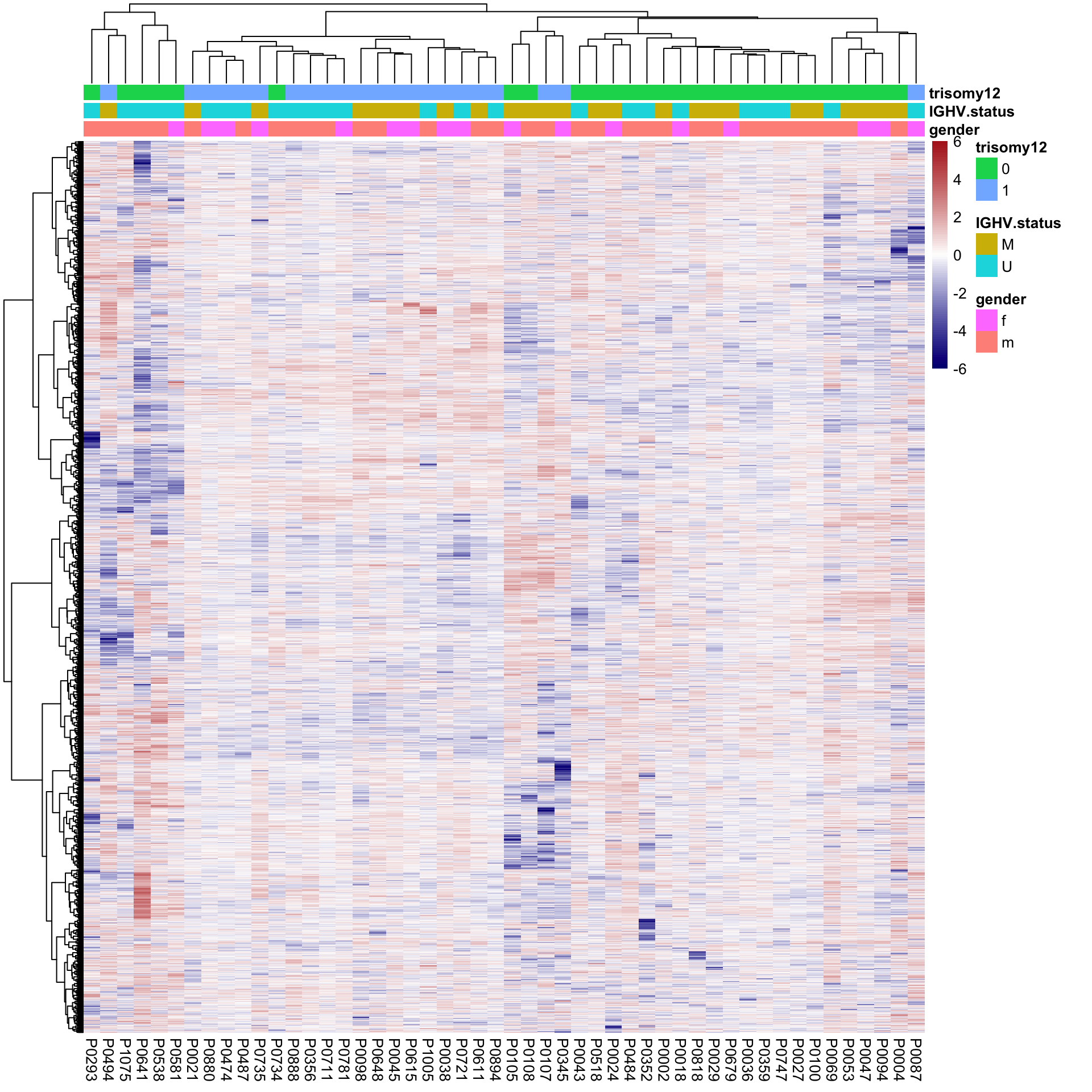 Better separation for trisomy12 can be observed
Better separation for trisomy12 can be observed
PCA using reconstructed matrix
pcOut <- prcomp(t(plotMat), center =TRUE, scale. = FALSE)
pcRes.new <- pcOut$x
eigs <- pcOut$sdev^2
varExp <- structure(eigs/sum(eigs),names = colnames(pcRes.new))
plotTab <- pcRes.new[,1:2] %>% data.frame() %>% cbind(colAnno[rownames(.),]) %>%
rownames_to_column("patID") %>% as_tibble()
ggplot(plotTab, aes(x=PC1, y=PC2, col = IGHV.status, shape = trisomy12)) + geom_point(size=4) +
xlab(sprintf("PC1 (%1.2f%%)",varExp[["PC1"]]*100)) +
ylab(sprintf("PC2 (%1.2f%%)",varExp[["PC2"]]*100))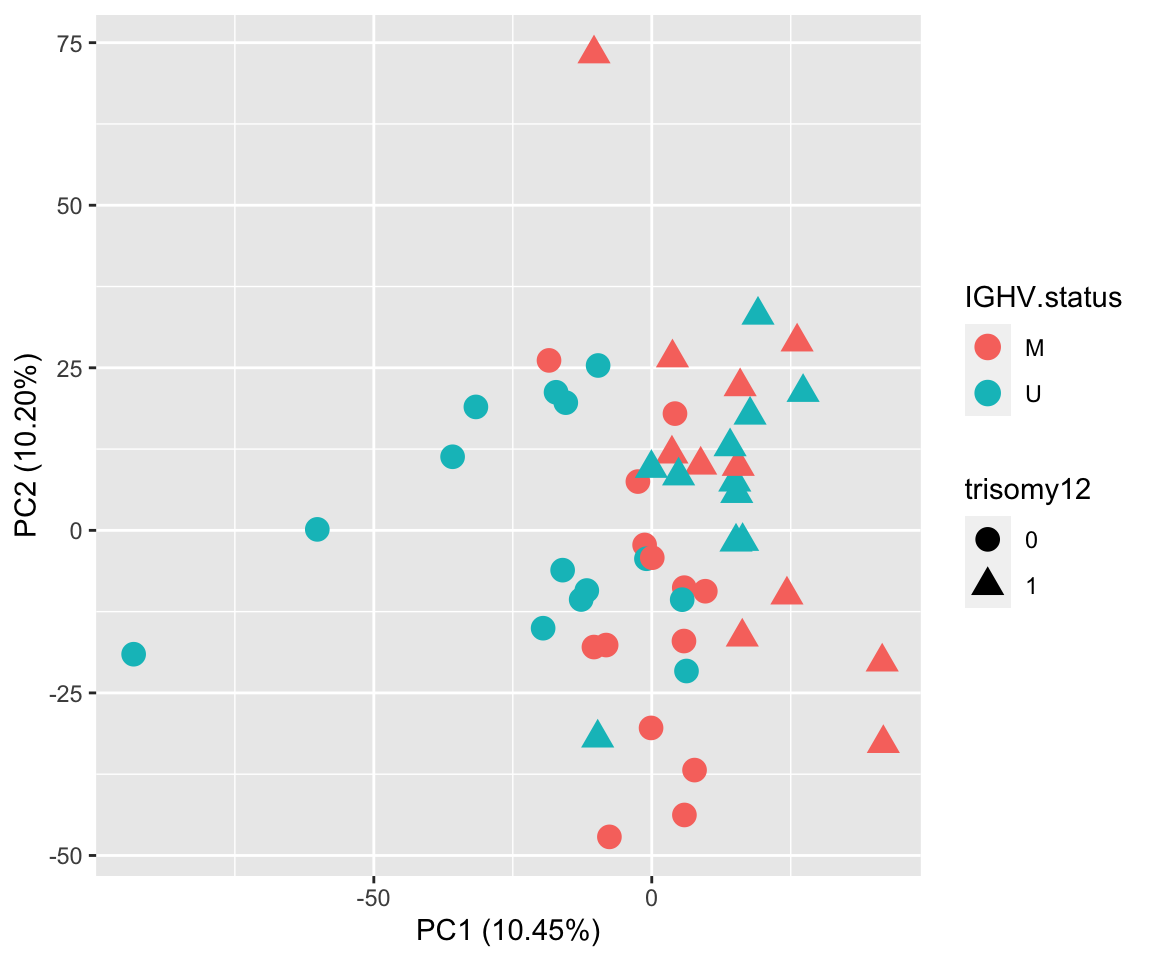 Some outliers dominate the variance
Some outliers dominate the variance
Save the post process object
assays(protCLL)[["QRILC_re"]] <- protMat.new
protCLL$PC1 <- pcRes[colnames(protCLL),1]
protCLL$PC2 <- pcRes[colnames(protCLL),2]
save(protCLL, file = "../output/timsTOF_processed.RData")timsTOF has some quality issues, and it's not very clear what the major variance in this dataset represents.
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] DBI_1.1.0 forcats_0.5.0
[3] stringr_1.4.0 dplyr_1.0.0
[5] purrr_0.3.4 readr_1.3.1
[7] tidyr_1.1.0 tibble_3.0.3
[9] ggplot2_3.3.2 tidyverse_1.3.0
[11] DESeq2_1.26.0 SummarizedExperiment_1.16.1
[13] DelayedArray_0.12.3 BiocParallel_1.20.1
[15] matrixStats_0.56.0 Biobase_2.46.0
[17] GenomicRanges_1.38.0 GenomeInfoDb_1.22.1
[19] IRanges_2.20.2 S4Vectors_0.24.4
[21] BiocGenerics_0.32.0 jyluMisc_0.1.5
[23] pheatmap_1.0.12 piano_2.2.0
[25] cowplot_1.0.0 limma_3.42.2
loaded via a namespace (and not attached):
[1] utf8_1.1.4 shinydashboard_0.7.1 tidyselect_1.1.0
[4] RSQLite_2.2.0 AnnotationDbi_1.48.0 htmlwidgets_1.5.1
[7] grid_3.6.0 maxstat_0.7-25 munsell_0.5.0
[10] codetools_0.2-16 DT_0.14 withr_2.2.0
[13] colorspace_1.4-1 knitr_1.29 rstudioapi_0.11
[16] ggsignif_0.6.0 labeling_0.3 git2r_0.27.1
[19] slam_0.1-47 GenomeInfoDbData_1.2.2 KMsurv_0.1-5
[22] bit64_0.9-7 farver_2.0.3 rprojroot_1.3-2
[25] vctrs_0.3.1 generics_0.0.2 TH.data_1.0-10
[28] xfun_0.15 sets_1.0-18 R6_2.4.1
[31] locfit_1.5-9.4 bitops_1.0-6 fgsea_1.12.0
[34] assertthat_0.2.1 promises_1.1.1 scales_1.1.1
[37] multcomp_1.4-13 nnet_7.3-14 gtable_0.3.0
[40] sandwich_2.5-1 workflowr_1.6.2 rlang_0.4.7
[43] genefilter_1.68.0 splines_3.6.0 rstatix_0.6.0
[46] acepack_1.4.1 broom_0.7.0 checkmate_2.0.0
[49] yaml_2.2.1 abind_1.4-5 modelr_0.1.8
[52] crosstalk_1.1.0.1 backports_1.1.8 httpuv_1.5.4
[55] Hmisc_4.4-0 tools_3.6.0 relations_0.6-9
[58] RPostgreSQL_0.6-2 ellipsis_0.3.1 gplots_3.0.4
[61] RColorBrewer_1.1-2 Rcpp_1.0.5 base64enc_0.1-3
[64] visNetwork_2.0.9 zlibbioc_1.32.0 RCurl_1.98-1.2
[67] ggpubr_0.4.0 rpart_4.1-15 zoo_1.8-8
[70] ggrepel_0.8.2 haven_2.3.1 cluster_2.1.0
[73] exactRankTests_0.8-31 fs_1.4.2 magrittr_1.5
[76] data.table_1.12.8 openxlsx_4.1.5 reprex_0.3.0
[79] survminer_0.4.7 mvtnorm_1.1-1 hms_0.5.3
[82] shinyjs_1.1 mime_0.9 evaluate_0.14
[85] xtable_1.8-4 XML_3.98-1.20 rio_0.5.16
[88] jpeg_0.1-8.1 readxl_1.3.1 gridExtra_2.3
[91] compiler_3.6.0 KernSmooth_2.23-17 crayon_1.3.4
[94] htmltools_0.5.0 mgcv_1.8-31 later_1.1.0.1
[97] Formula_1.2-3 geneplotter_1.64.0 lubridate_1.7.9
[100] dbplyr_1.4.4 MASS_7.3-51.6 Matrix_1.2-18
[103] car_3.0-8 cli_2.0.2 marray_1.64.0
[106] gdata_2.18.0 igraph_1.2.5 pkgconfig_2.0.3
[109] km.ci_0.5-2 foreign_0.8-71 xml2_1.3.2
[112] annotate_1.64.0 XVector_0.26.0 drc_3.0-1
[115] rvest_0.3.5 digest_0.6.25 rmarkdown_2.3
[118] cellranger_1.1.0 fastmatch_1.1-0 survMisc_0.5.5
[121] htmlTable_2.0.1 curl_4.3 shiny_1.5.0
[124] gtools_3.8.2 lifecycle_0.2.0 nlme_3.1-148
[127] jsonlite_1.7.0 carData_3.0-4 fansi_0.4.1
[130] pillar_1.4.6 lattice_0.20-41 fastmap_1.0.1
[133] httr_1.4.1 plotrix_3.7-8 survival_3.2-3
[136] glue_1.4.1 zip_2.0.4 png_0.1-7
[139] bit_1.1-15.2 stringi_1.4.6 blob_1.2.1
[142] latticeExtra_0.6-29 caTools_1.18.0 memoise_1.1.0