Differential methylated region (DMR) analysis for targeted gene list
Junyan Lu
Last updated: 2023-09-04
Checks: 5 1
Knit directory: RA_Tcell_omics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221110) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the
results is critical for reproducibility. To start using Git, open the
Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full
reproducibility benefits of using workflowr, please see
?wflow_start.
Load libraries
Global variables
DMR analysis to identify significant DMRs that contain specific regions associated with genes (use hg19 annotation)
Preprocessing
Load data
Get unique symbol
Get interested gene list
Process methylation dataset
[1] 20884 23Differential methylation analysis on probe level
SVA to identify unknown confounder
Number of significant surrogate variables is: 3
Iteration (out of 5 ):1 2 3 4 5 DE test using limma
Using dmrff to identify DMRs
[dmrff.candidates] Mon Sep 4 15:16:22 2023 Found 1087 candidate regions. Annotate MDRs
Save significant DMRs with annotation
symbol DMR site chr start end number_CpG estimate
1 SLC16A11 dmr1 Body chr17 6945510 6946086 4 0.9815272
2 CACNG2 dmr2 TSS200 chr22 37099095 37099785 7 0.6132989
3 GDA dmr3 5'UTR chr9 74764261 74764263 2 1.0916481
4 RGN dmr4 TSS1500 chrX 46937571 46938148 7 0.9524670
5 NOS1 dmr5 1stExon chr12 117799370 117799749 3 0.6260553
6 CYP1B1 dmr6 Body chr2 38300537 38300885 4 1.0469769
p.value p.adjust
1 8.640059e-15 1.981943e-10
2 1.004762e-07 2.304823e-03
3 1.316342e-07 3.019557e-03
4 2.047986e-07 4.697875e-03
5 2.320634e-07 5.323303e-03
6 1.351948e-06 3.101233e-02Download xlsx table: DMR_regions.xlsx
Visuaize enhancer/promoter methylation status per gene
Download plots in zip file: plot_DMR.zip
DMR analysis to identify significant DMRs that contain GeneHancer regions
Process enhancer list
Create genomic ranges object
Find GC probs in the enhancer region
Perform differential analysis
Process methylation dataset
[1] 85102 23SVA to identify unknwon confounder
Number of significant surrogate variables is: 3
Iteration (out of 5 ):1 2 3 4 5 DE test using limma
Add mean difference of beta values
Get enhancer/promoter regions that contains differentially expressed probes
Comine the two tables
Using dmrff to identify DMRs
[dmrff.candidates] Mon Sep 4 15:17:16 2023 Found 4660 candidate regions. Annotate DMRs
gene enhancerId feature chr start end number_CpG
1 SLC16A11 GH17J007041 Promoter/Enhancer chr17 6945510 6946086 4
1.1 SLC16A13 GH17J007041 Promoter/Enhancer chr17 6945510 6946086 4
2 DDC GH07J050790 Promoter/Enhancer chr7 50861467 50861750 11
3 USP8 GH15J050181 Promoter/Enhancer chr15 50473854 50474221 6
3.1 HDC GH15J050181 Promoter/Enhancer chr15 50473854 50474221 6
4 GART GH21J033400 Promoter/Enhancer chr21 34775001 34775045 5
estimate p.value p.adjust
1 0.9974066 2.710935e-15 2.566333e-10
1.1 0.9974066 2.710935e-15 2.566333e-10
2 0.8361415 5.662111e-13 5.360094e-08
3 0.8601667 1.795356e-11 1.699592e-06
3.1 0.8601667 1.795356e-11 1.699592e-06
4 1.6298080 4.465063e-10 4.226896e-05Download xlsx table: DMR_GeneHancer.xlsx
Visuaize enhancer/promoter methylation status per gene
Download plots in zip file: plot_geneEnhancer.zip
Heatmap visualization of all significant DMRs
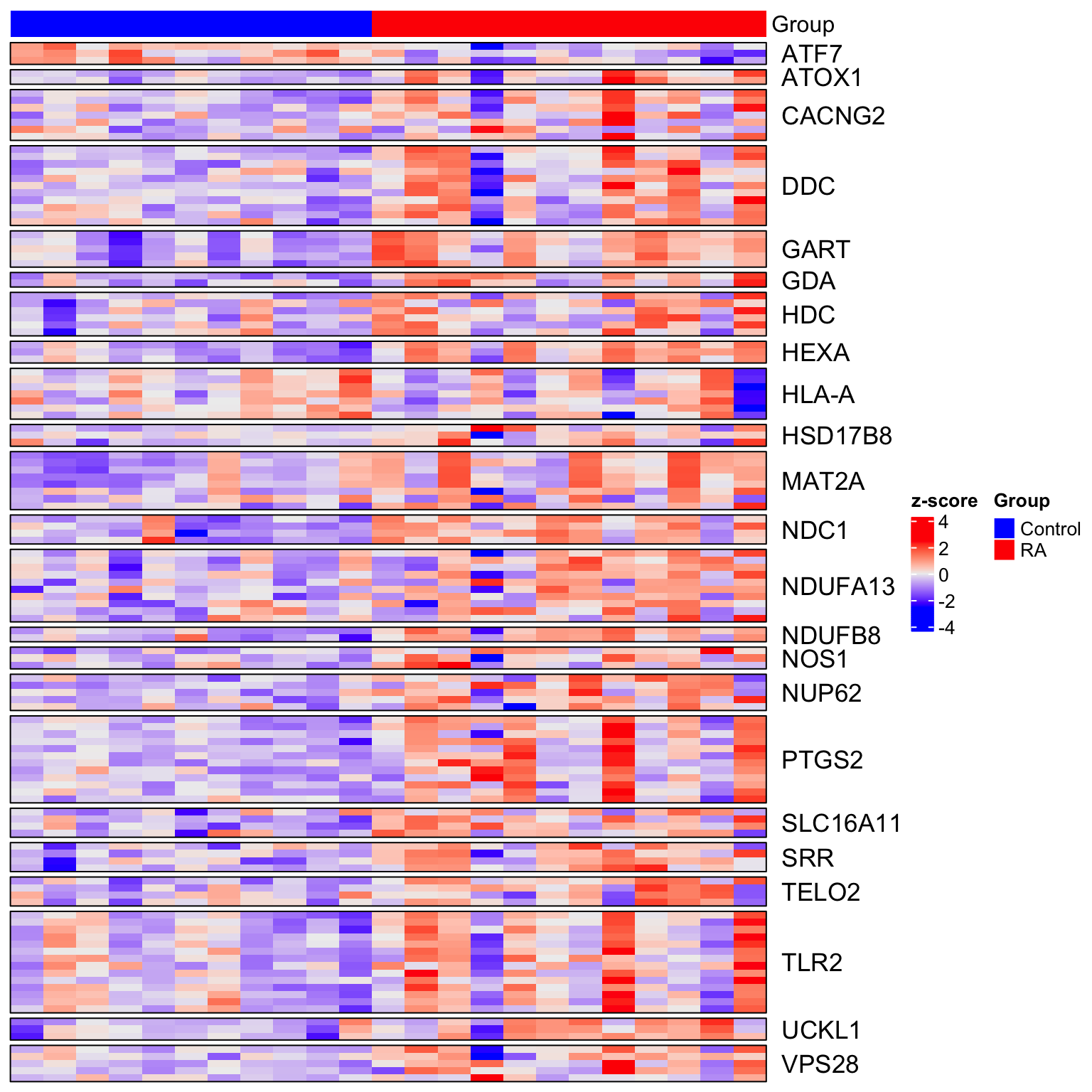
Example plot of enhancers
pdf
2 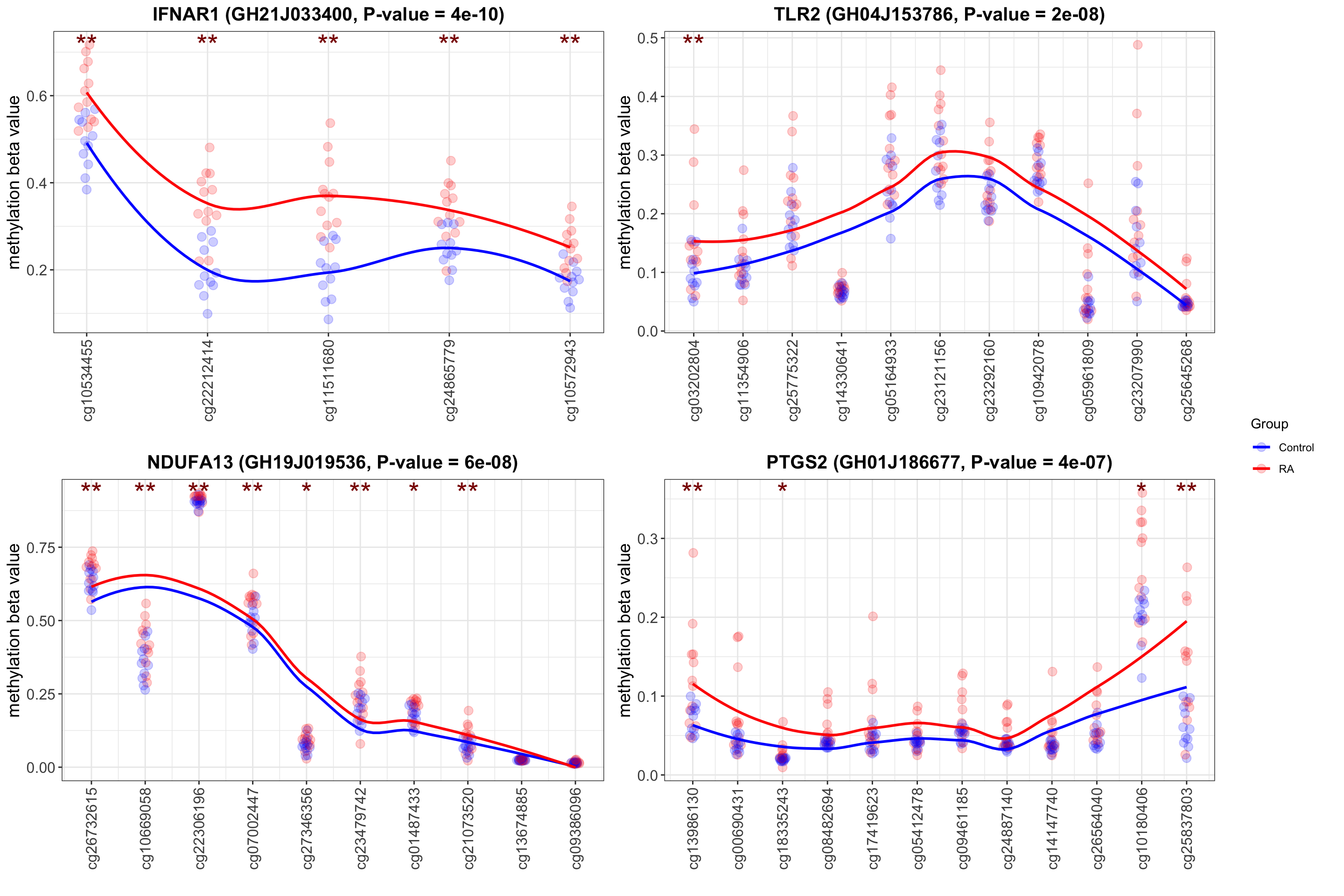
R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] cowplot_1.1.1 ComplexHeatmap_2.12.1
[3] gridExtra_2.3 sva_3.44.0
[5] BiocParallel_1.30.3 genefilter_1.78.0
[7] mgcv_1.8-40 nlme_3.1-158
[9] forcats_0.5.1 stringr_1.4.1
[11] dplyr_1.0.9 purrr_0.3.4
[13] readr_2.1.2 tidyr_1.2.0
[15] tibble_3.1.8 ggplot2_3.4.1
[17] tidyverse_1.3.2 pheatmap_1.0.12
[19] SummarizedExperiment_1.26.1 Biobase_2.56.0
[21] GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[23] IRanges_2.30.0 S4Vectors_0.34.0
[25] BiocGenerics_0.42.0 MatrixGenerics_1.8.1
[27] matrixStats_0.62.0 limma_3.52.2
loaded via a namespace (and not attached):
[1] utf8_1.2.2 shinydashboard_0.7.2 tidyselect_1.1.2
[4] RSQLite_2.2.15 AnnotationDbi_1.58.0 htmlwidgets_1.5.4
[7] maxstat_0.7-25 munsell_0.5.0 ragg_1.2.2
[10] codetools_0.2-18 DT_0.23 withr_2.5.0
[13] colorspace_2.0-3 highr_0.9 knitr_1.39
[16] rstudioapi_0.13 ggsignif_0.6.3 labeling_0.4.2
[19] git2r_0.30.1 slam_0.1-50 GenomeInfoDbData_1.2.8
[22] KMsurv_0.1-5 bit64_4.0.5 farver_2.1.1
[25] rprojroot_2.0.3 vctrs_0.5.2 generics_0.1.3
[28] TH.data_1.1-1 xfun_0.31 sets_1.0-21
[31] R6_2.5.1 doParallel_1.0.17 ggbeeswarm_0.6.0
[34] clue_0.3-61 locfit_1.5-9.6 bitops_1.0-7
[37] cachem_1.0.6 fgsea_1.22.0 DelayedArray_0.22.0
[40] assertthat_0.2.1 promises_1.2.0.1 scales_1.2.0
[43] multcomp_1.4-19 googlesheets4_1.0.0 beeswarm_0.4.0
[46] gtable_0.3.0 Cairo_1.6-0 dmrff_1.1.0
[49] sandwich_3.0-2 workflowr_1.7.0 rlang_1.0.6
[52] systemfonts_1.0.4 GlobalOptions_0.1.2 splines_4.2.0
[55] rstatix_0.7.0 gargle_1.2.0 broom_1.0.0
[58] yaml_2.3.5 abind_1.4-5 modelr_0.1.8
[61] backports_1.4.1 httpuv_1.6.6 tools_4.2.0
[64] relations_0.6-12 ellipsis_0.3.2 gplots_3.1.3
[67] jquerylib_0.1.4 RColorBrewer_1.1-3 Rcpp_1.0.9
[70] visNetwork_2.1.0 zlibbioc_1.42.0 RCurl_1.98-1.7
[73] ggpubr_0.4.0 GetoptLong_1.0.5 zoo_1.8-10
[76] haven_2.5.0 cluster_2.1.3 exactRankTests_0.8-35
[79] fs_1.5.2 magrittr_2.0.3 magick_2.7.3
[82] data.table_1.14.8 circlize_0.4.15 reprex_2.0.1
[85] survminer_0.4.9 googledrive_2.0.0 mvtnorm_1.1-3
[88] hms_1.1.1 shinyjs_2.1.0 mime_0.12
[91] evaluate_0.15 xtable_1.8-4 XML_3.99-0.10
[94] readxl_1.4.0 shape_1.4.6 compiler_4.2.0
[97] writexl_1.4.0 KernSmooth_2.23-20 crayon_1.5.2
[100] htmltools_0.5.4 later_1.3.0 tzdb_0.3.0
[103] lubridate_1.8.0 DBI_1.1.3 dbplyr_2.2.1
[106] MASS_7.3-58 jyluMisc_0.1.5 Matrix_1.5-4
[109] car_3.1-0 cli_3.4.1 marray_1.74.0
[112] parallel_4.2.0 igraph_1.3.4 pkgconfig_2.0.3
[115] km.ci_0.5-6 piano_2.12.0 xml2_1.3.3
[118] foreach_1.5.2 annotate_1.74.0 vipor_0.4.5
[121] bslib_0.4.1 XVector_0.36.0 drc_3.0-1
[124] rvest_1.0.2 digest_0.6.30 Biostrings_2.64.0
[127] rmarkdown_2.14 cellranger_1.1.0 fastmatch_1.1-3
[130] survMisc_0.5.6 edgeR_3.38.1 shiny_1.7.4
[133] gtools_3.9.3 rjson_0.2.21 lifecycle_1.0.3
[136] jsonlite_1.8.3 carData_3.0-5 fansi_1.0.3
[139] pillar_1.8.0 lattice_0.20-45 KEGGREST_1.36.3
[142] fastmap_1.1.0 httr_1.4.3 plotrix_3.8-2
[145] survival_3.4-0 glue_1.6.2 png_0.1-7
[148] iterators_1.0.14 bit_4.0.4 stringi_1.7.8
[151] sass_0.4.2 blob_1.2.3 textshaping_0.3.6
[154] caTools_1.18.2 memoise_2.0.1