Exploratory and Power Analysis of the RA metabolomic and proteomic data
Junyan Lu
Last updated: 2023-03-15
Checks: 5 1
Knit directory: RA_Tcell_omics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221110) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the
results is critical for reproducibility. To start using Git, open the
Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full
reproducibility benefits of using workflowr, please see
?wflow_start.
Load libraries
library(vsn)
library(pwr)
library(jyluMisc)
library(SummarizedExperiment)
library(tidyverse)
knitr::opts_chunk$set(warning = FALSE, message = FALSE)Analysis of metabolimic data
Preprocessing
metaTab <- readxl::read_xlsx("../data/Metabolomics-Exvivo_CD8.xlsx") %>%
pivot_longer(-c("Sample name","Group"), values_to = "value", names_to = "metabolite") %>%
dplyr::rename(sampleName = `Sample name`)
metaMat <- metaTab %>% select(-Group) %>%
pivot_wider(names_from = "sampleName", values_from = "value") %>%
column_to_rownames("metabolite") %>%
as.matrix()Visualize overall data distribution
Per sample
ggplot(metaTab, aes(x=sampleName, y=value)) +
geom_boxplot() + geom_point(aes(col = Group)) +
theme(axis.text = element_text(angle = 90, hjust = 1, vjust = 0.5))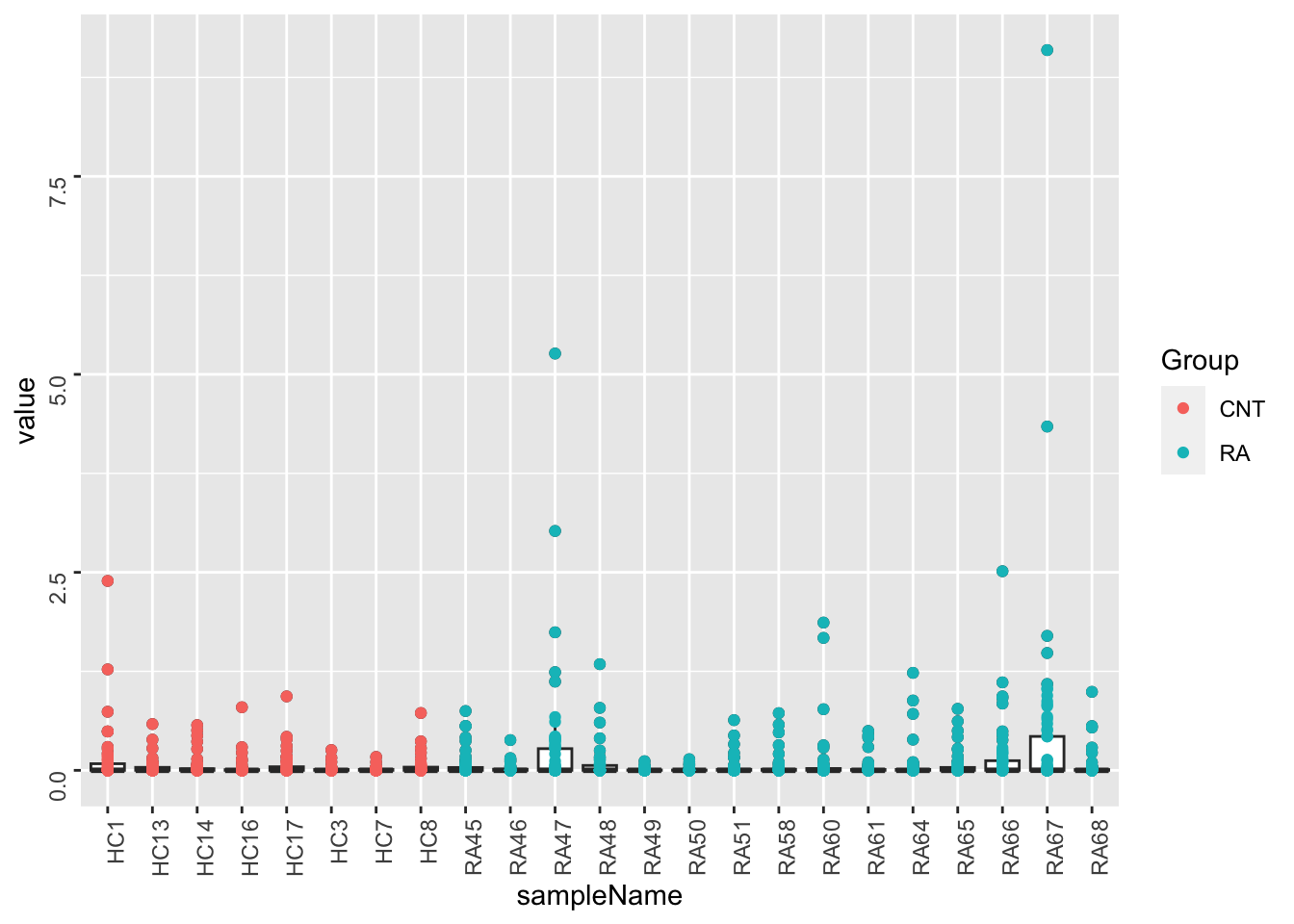
Per metabolite
ggplot(metaTab, aes(x=metabolite, y=value)) +
geom_boxplot() + geom_point(aes(col = Group)) +
theme(axis.text = element_text(angle = 90, hjust = 1, vjust = 0.5))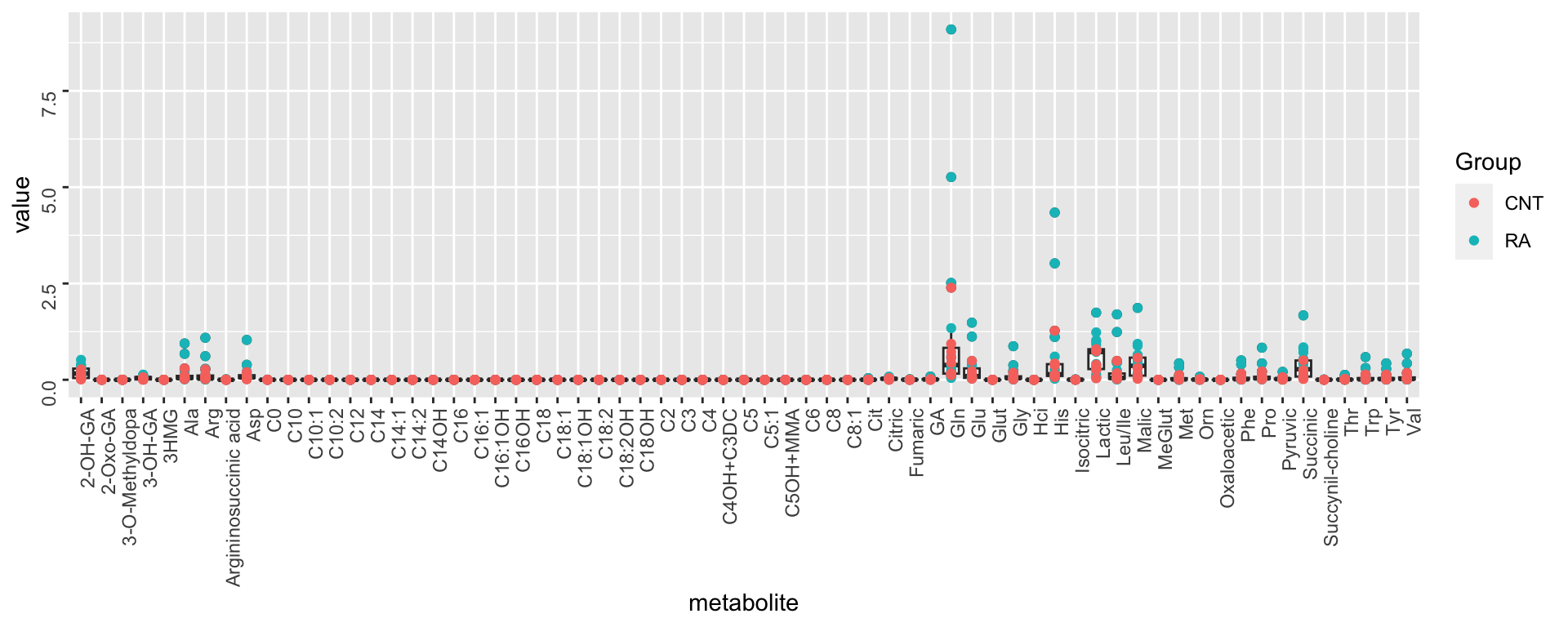 The abundance of different metabolites are very different.
Transformation and Normalization may not be needed actually
The abundance of different metabolites are very different.
Transformation and Normalization may not be needed actually
PCA
pcRes <- prcomp(t(metaMat), scale. = TRUE, center = TRUE)$x
plotTab <- as_tibble(pcRes, rownames = "sampleName") %>%
mutate(Group = metaTab[match(sampleName, metaTab$sampleName),]$Group)
ggplot(plotTab, aes(x=PC1, y=PC2, col = Group)) +
geom_point()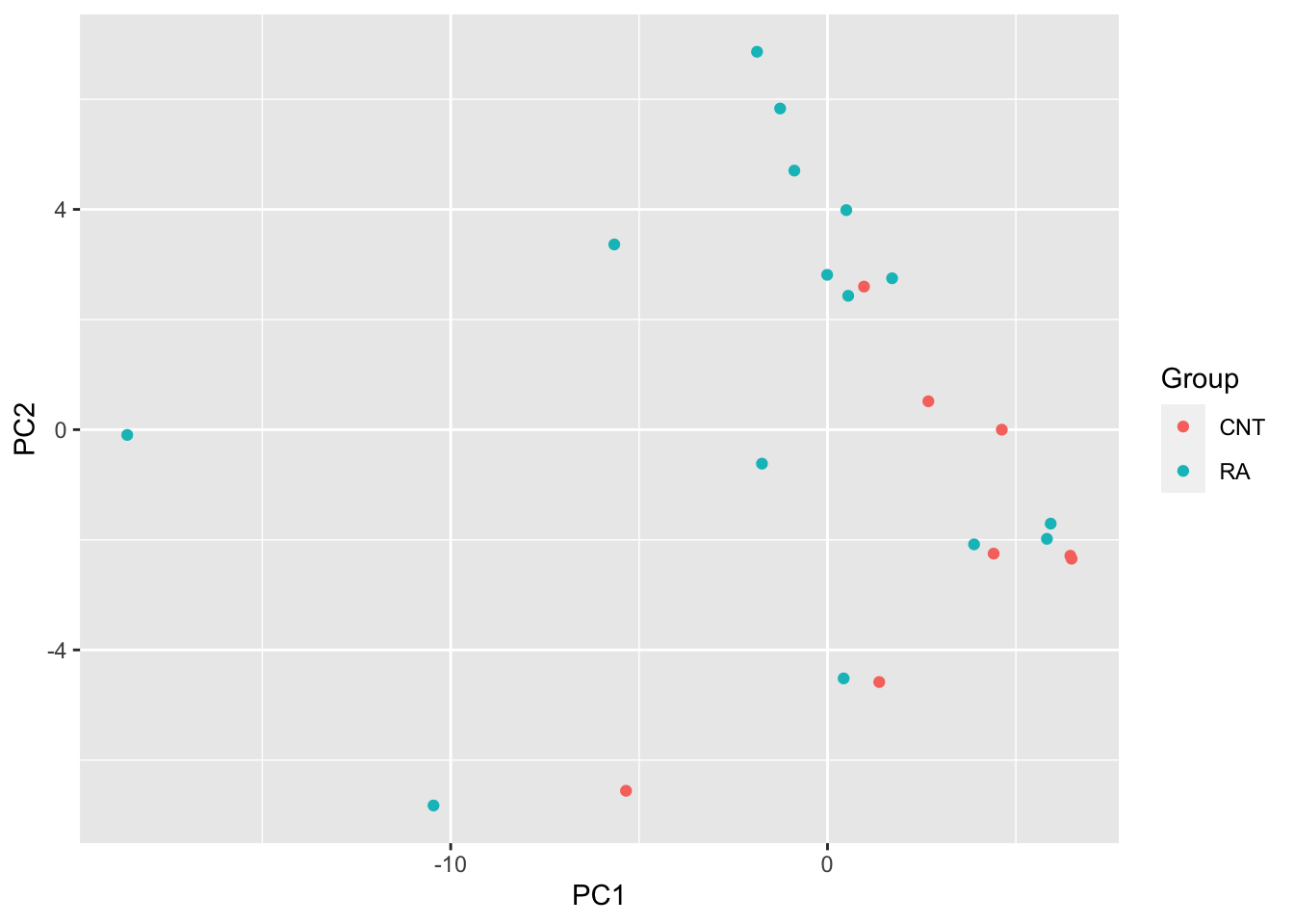
Differential abundance test
library(limma)
group <- factor(metaTab[match(colnames(metaMat), metaTab$sampleName),]$Group)
designMat <- model.matrix(~group)
lmFit <- lmFit(metaMat, design = designMat)
fit2 <- eBayes(lmFit)
resTab <- topTable(fit2, number = Inf) %>%
as_tibble(rownames = "metabolite")
hist(resTab$P.Value)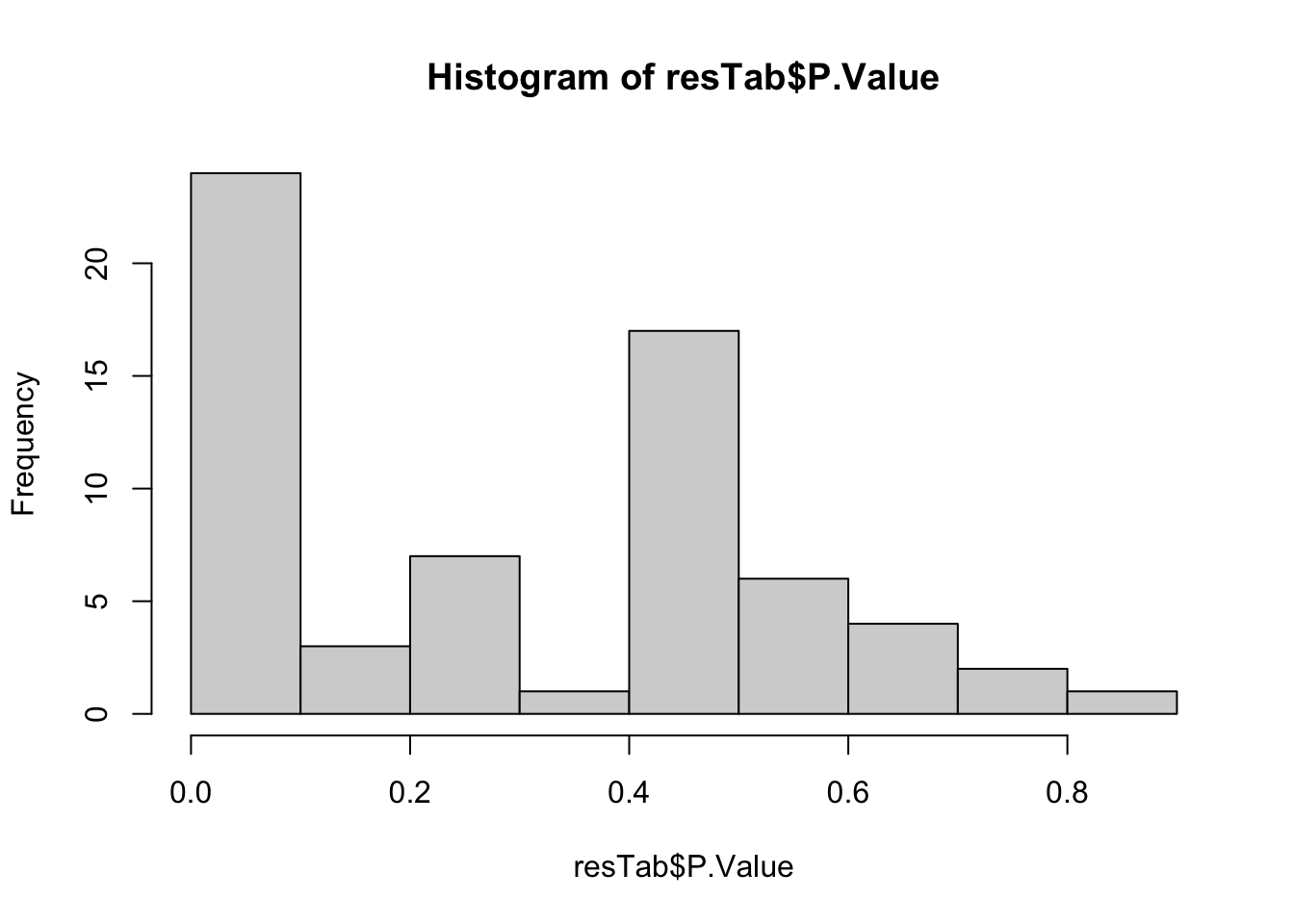
Plot significant associations
pList <- lapply(seq(nrow(filter(resTab, P.Value <= 0.05))), function(i) {
rec <- resTab[i,]
plotTab <- filter(metaTab, metabolite == rec$metabolite)
ggplot(plotTab, aes(x=Group, y=value)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = Group), size=3) +
ggtitle(sprintf("%s (P=%s)",rec$metabolite,formatC(rec$P.Value,digits = 2))) +
theme_bw() +
theme(legend.position = "none")
})
cowplot::plot_grid(plotlist = pList,ncol=3)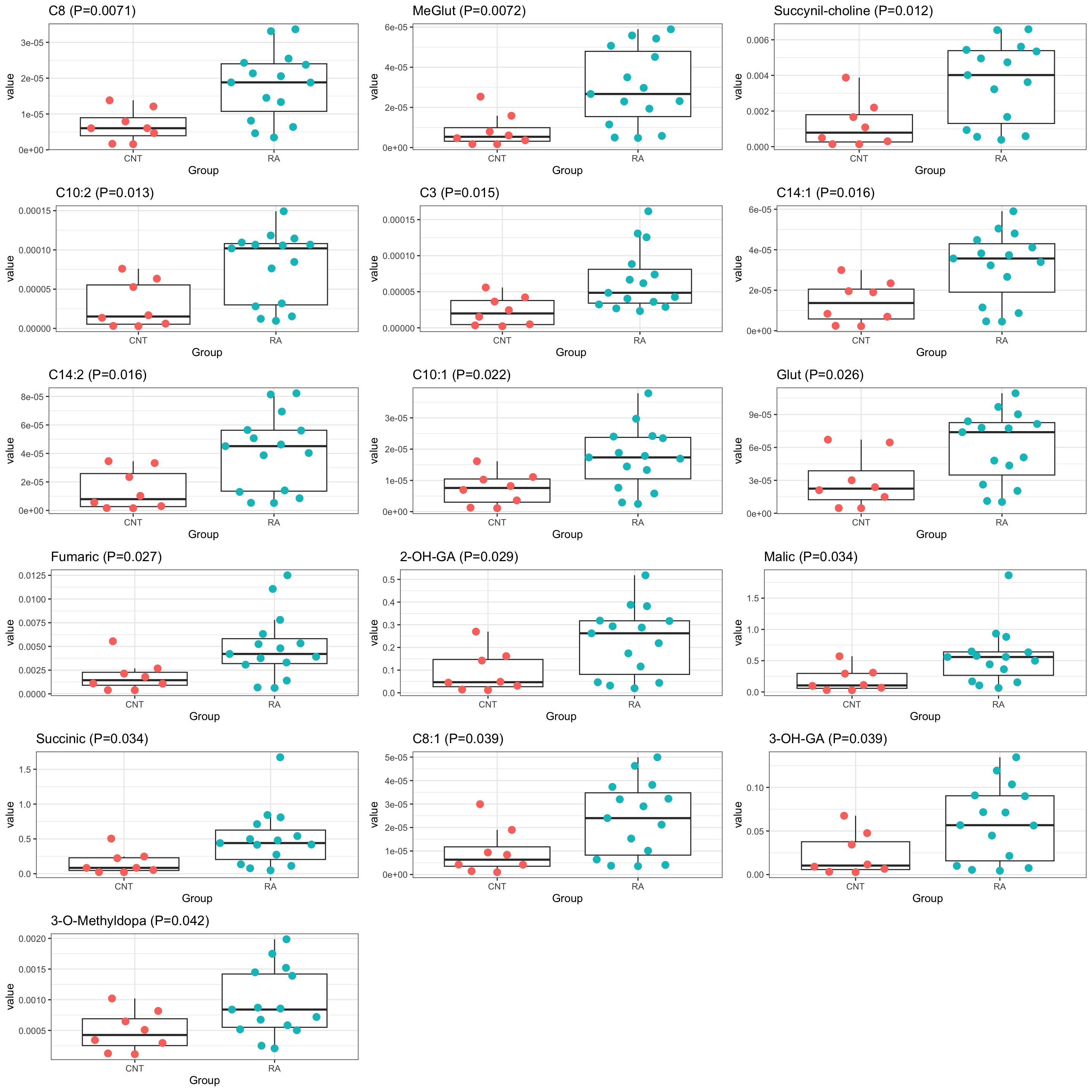
Power analysis to estimate sample size
meanTab <- metaTab %>%
group_by(Group, metabolite) %>%
summarise(mean = mean(value)) %>%
pivot_wider(names_from = Group, values_from = mean)
sdTab <- metaTab %>%
group_by(metabolite) %>%
summarise(sd = sd(value))
dTab <- left_join(meanTab, sdTab) %>%
mutate(d= abs(RA-CNT)/sd)
nTab <- lapply(seq(nrow(dTab)), function(i) {
rec <- dTab[i,]
n1 <- pwr.t.test(d = rec$d , sig.level = 0.05,
power = 0.9, alternative = "two.sided")$n
n2 <- pwr.t.test(d = rec$d , sig.level = 0.01,
power = 0.9, alternative = "two.sided")$n
tibble(metabolite = rec$metabolite,
n0.05 = as.integer(n1), n0.01=as.integer(n2))
}) %>% bind_rows() %>%
arrange(n0.05)Table of sample number at certain p-value threshold
nTab %>% DT::datatable()Bar-plot
plotTab <- nTab %>% mutate(metabolite = factor(metabolite, levels = metabolite)) %>%
pivot_longer(-metabolite)
ggplot(plotTab, aes(x=metabolite, y=value, fill = name)) +
geom_bar(stat = "identity", position = "dodge") +
scale_color_discrete(name = "P-value cut",
labels = c("0.01","0.05")) +
coord_cartesian(ylim=c(0,100)) +
theme_bw() +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
ylab("samples required per group")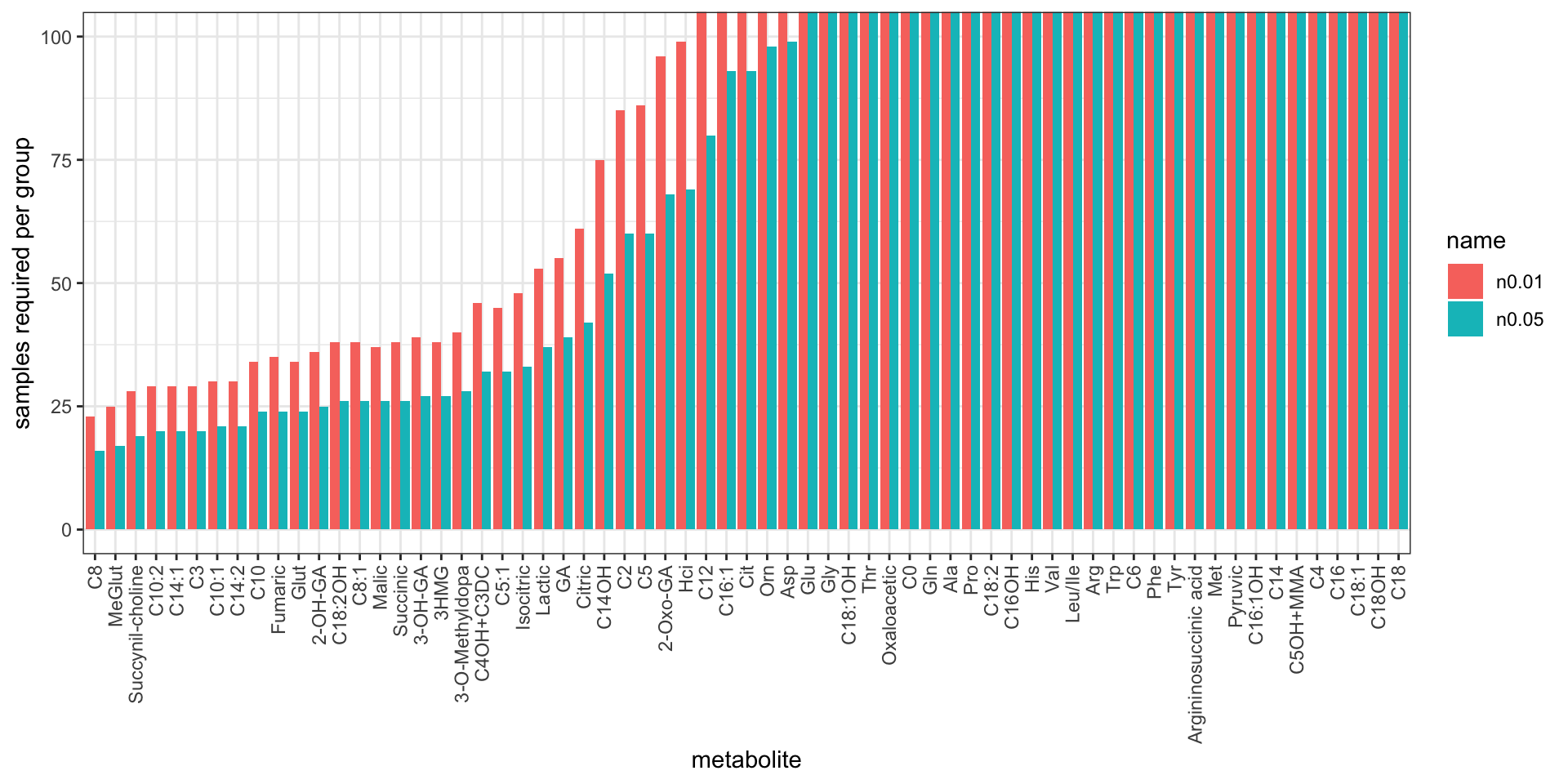
Analysis of the proteomic dataset
Data input
protData <- readxl::read_xlsx("../data/TF0489/TF0489_filtered_proteinGroups.xlsx") %>%
select(`Majority protein IDs`, `Gene names`, contains("LFQ intensity")) %>%
dplyr::rename(protID = `Majority protein IDs`, symbol = `Gene names`) %>%
pivot_longer(-c("protID","symbol")) %>%
mutate(name = str_remove(name,"LFQ intensity "))Annotate samples
patAnno <- readxl::read_xlsx("../data/TF0489/Sample-Information.xlsx") %>%
filter(!is.na(`Patient Group`))
patAnno <- patAnno[,c(1,2,4,5,6,7,8)]
colnames(patAnno) <- c("name","sampleName","Group","protConc","quantStart","sampleVol","bufferComp")
patAnno <- mutate(patAnno, name = paste0("Sample",sprintf("%02s",name)))
protData <- protData %>%
left_join(patAnno, by = "name") %>%
dplyr::rename(sampleID = name) %>%
dplyr::rename(name = symbol) %>%
mutate(ID = protID)Check DNMT1 expression before any transformation
dnmtTab <- filter(protData, name == "DNMT1")
ggplot(dnmtTab, aes(x=Group, y=value)) +
geom_boxplot() +
geom_point()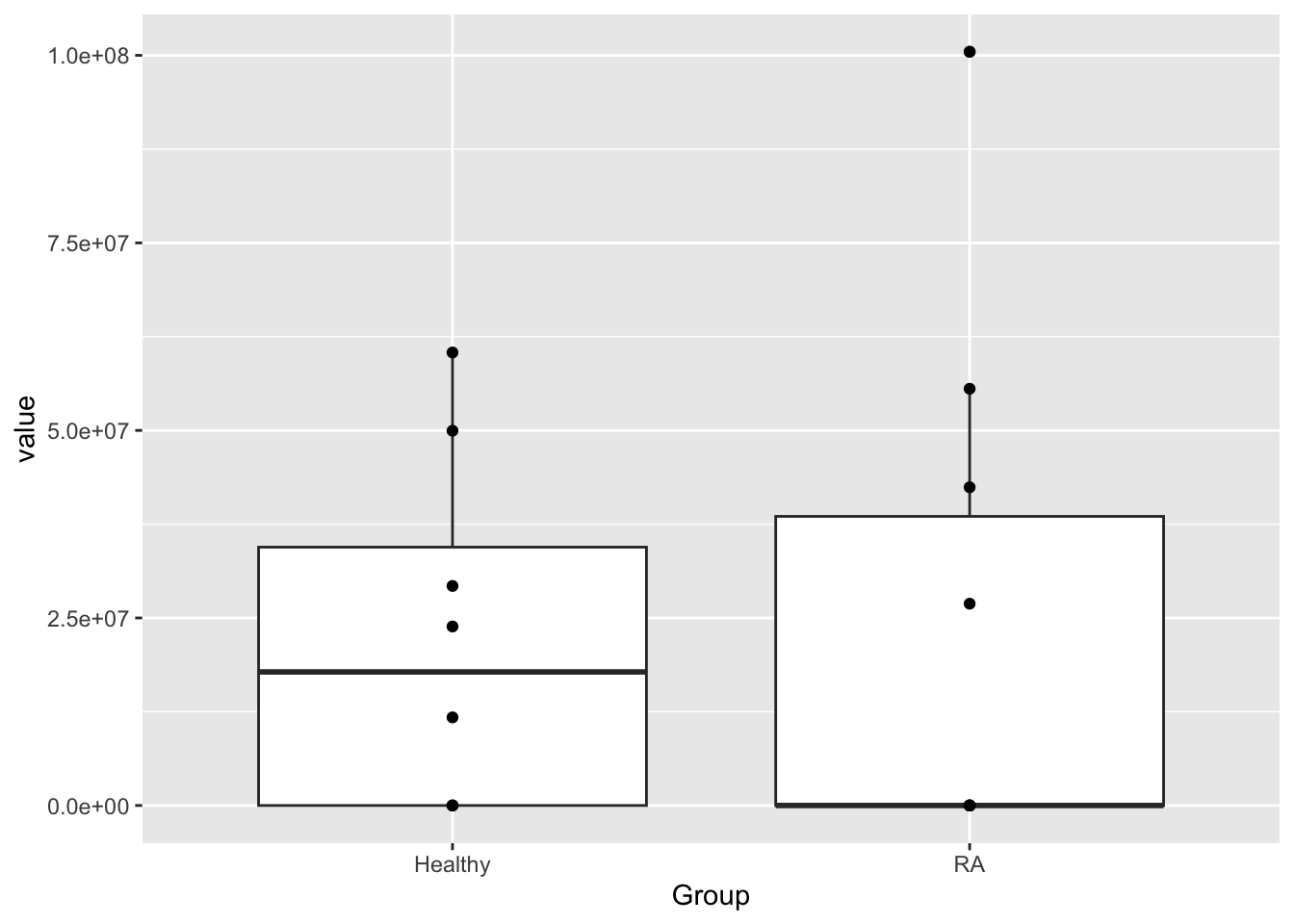
Remove undetected values
protData <- protData %>% filter(value >0)Created summarised experiment
protObj <- jyluMisc::tidyToSum(protData, "protID", "sampleID","value",
annoRow = c("name","ID"),
annoCol = c("sampleName","Group",
"protConc",
"sampleVol","bufferComp"))protMat <- assay(protObj)
boxplot(protMat) Needs proprocessing
Needs proprocessing
Preprocess proteomic data
Missing value per sample
countMat <- assay(protObj)
plotTab <- tibble(sample = colnames(protObj),
perNA = colSums(is.na(countMat))/nrow(countMat))
ggplot(plotTab, aes(x=sample, y=1-perNA)) +
geom_bar(stat = "identity") +
ylab("completeness") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust=0))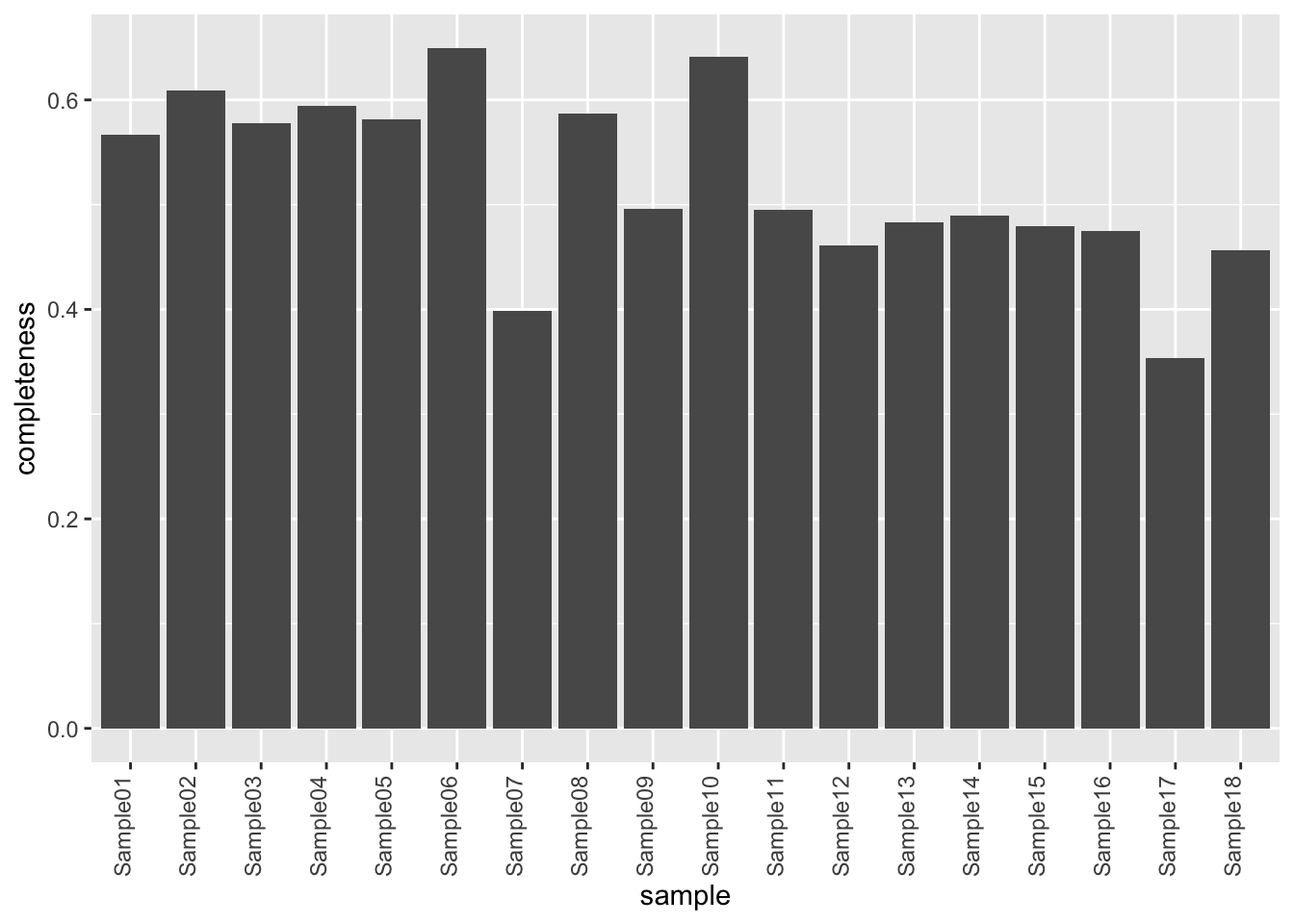
Plot a cumulative curve of missing value cut-off and remaining number of features
missRate <- tibble(id = rownames(countMat),
rate = rowSums(is.na(countMat))/ncol(countMat))
cumTab <- lapply(seq(0,1,0.05), function(cutRate) {
tibble(cut= cutRate,
per = sum(missRate$rate <= cutRate)/nrow(missRate))
} ) %>%
bind_rows()
ggplot(cumTab, aes(x=cut,y=per)) +
geom_line() +
xlab("Allowed missing value rate") +
ylab("Percentage of remaining features")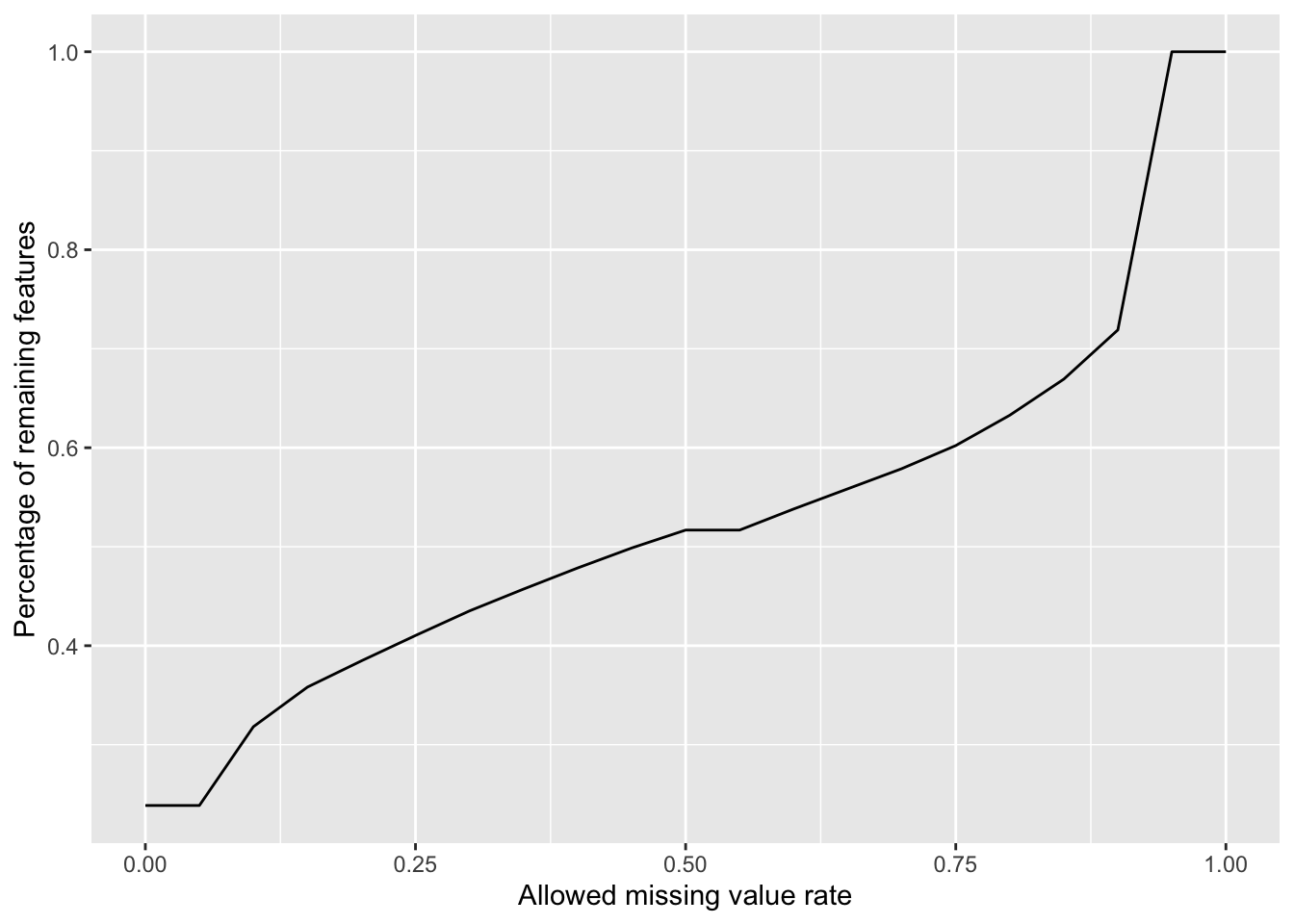 Missing value heatmap to check missing value structure
Missing value heatmap to check missing value structure
DEP::plot_missval(protObj)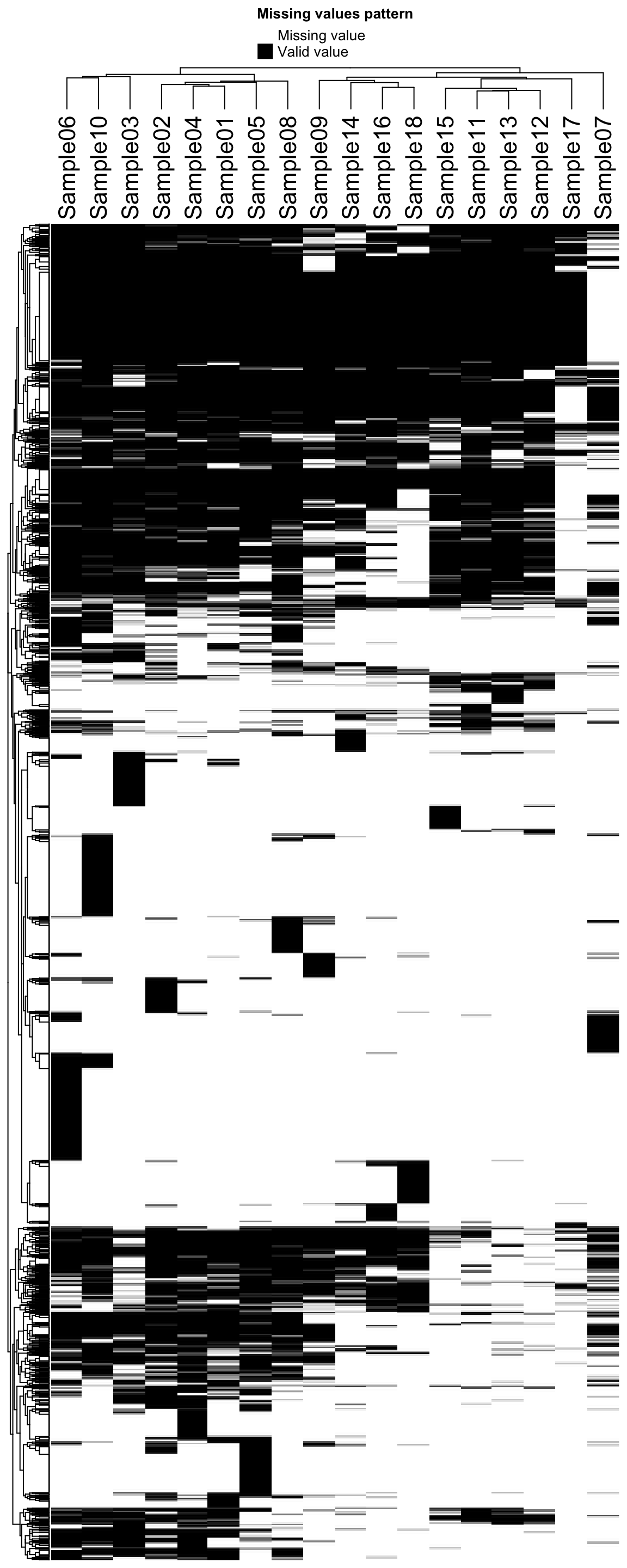 Rather random
Rather random
Keep proteins detected in at least half of the sample (missing rate <= 0.5)
protFilt <- protObj[filter(missRate, rate <=0.5)$id,]
dim(protFilt)[1] 2766 18Data distribution
countMat <- assay(protFilt)
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value)) %>%
mutate(log2Val = log2(value))ggplot(countTab, aes(x=name, y=log2Val)) +
geom_boxplot() + geom_point()
Imputation and normalization
Vst
protMat <- assay(protFilt)
fitVsn <- vsn::vsnMatrix(protMat)
normMat <- predict(fitVsn, newdata = protMat)
protNorm <- protFilt
assay(protNorm) <- normMatImputation
protImp <- DEP::impute(protNorm, "QRILC")
assays(protFilt)[["norm"]] <- normMat
assays(protFilt)[["imputed"]] <- assay(protImp)
rowData(protFilt) <- rowData(protImp)Distribution after normalizaiton
countMat <- assays(protFilt)[["imputed"]]
countTab <- countMat %>% as_tibble(rownames = "id") %>%
pivot_longer(-id) %>%
filter(!is.na(value))
ggplot(countTab, aes(x=name, y=value)) +
geom_boxplot() + geom_point()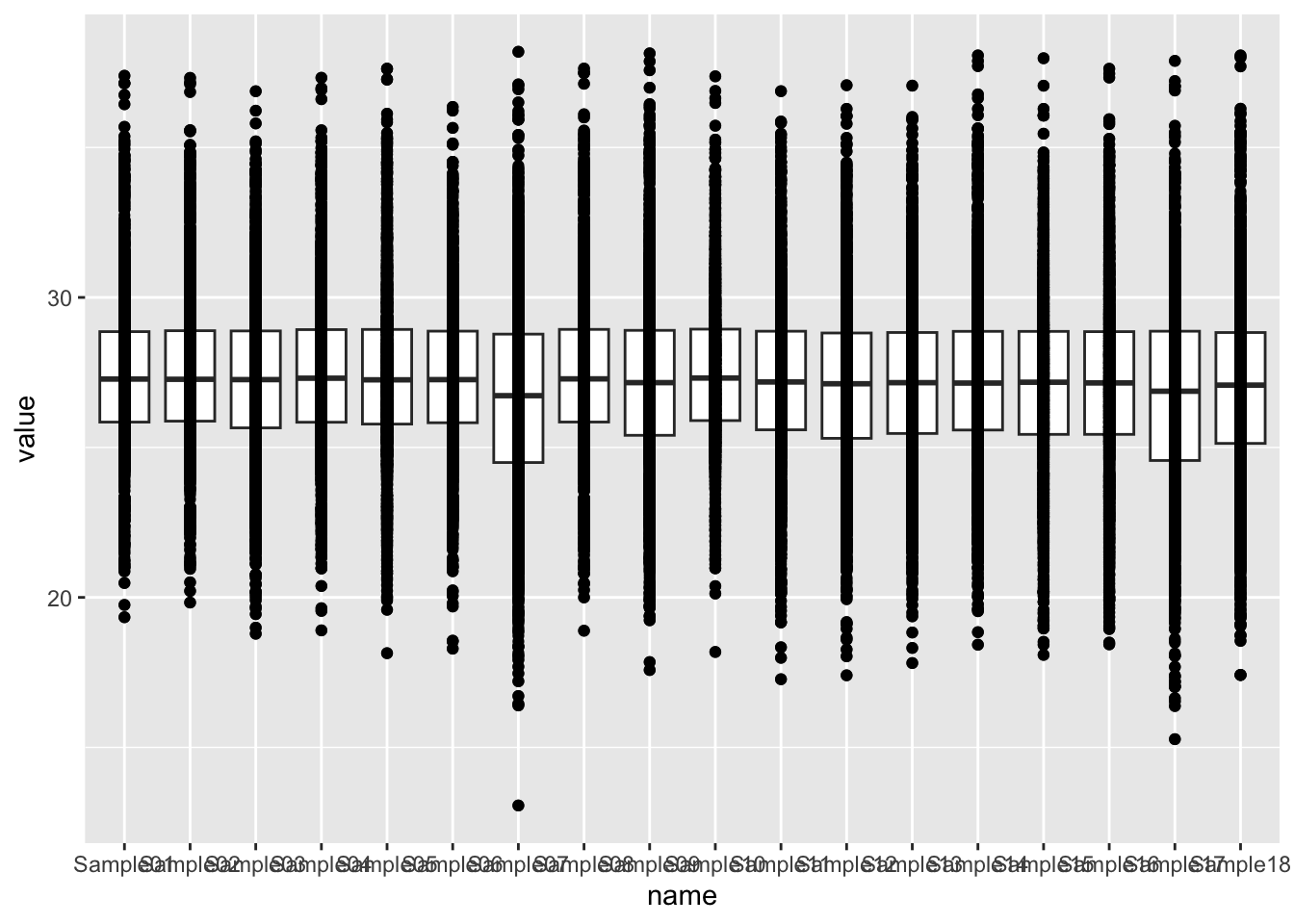 Mean versus variant plot
Mean versus variant plot
plotTab <- tibble(meanVal = rowMeans(countMat),
var = apply(countMat, 1, var))
ggplot(plotTab, aes(x=meanVal,y=var)) +
geom_point()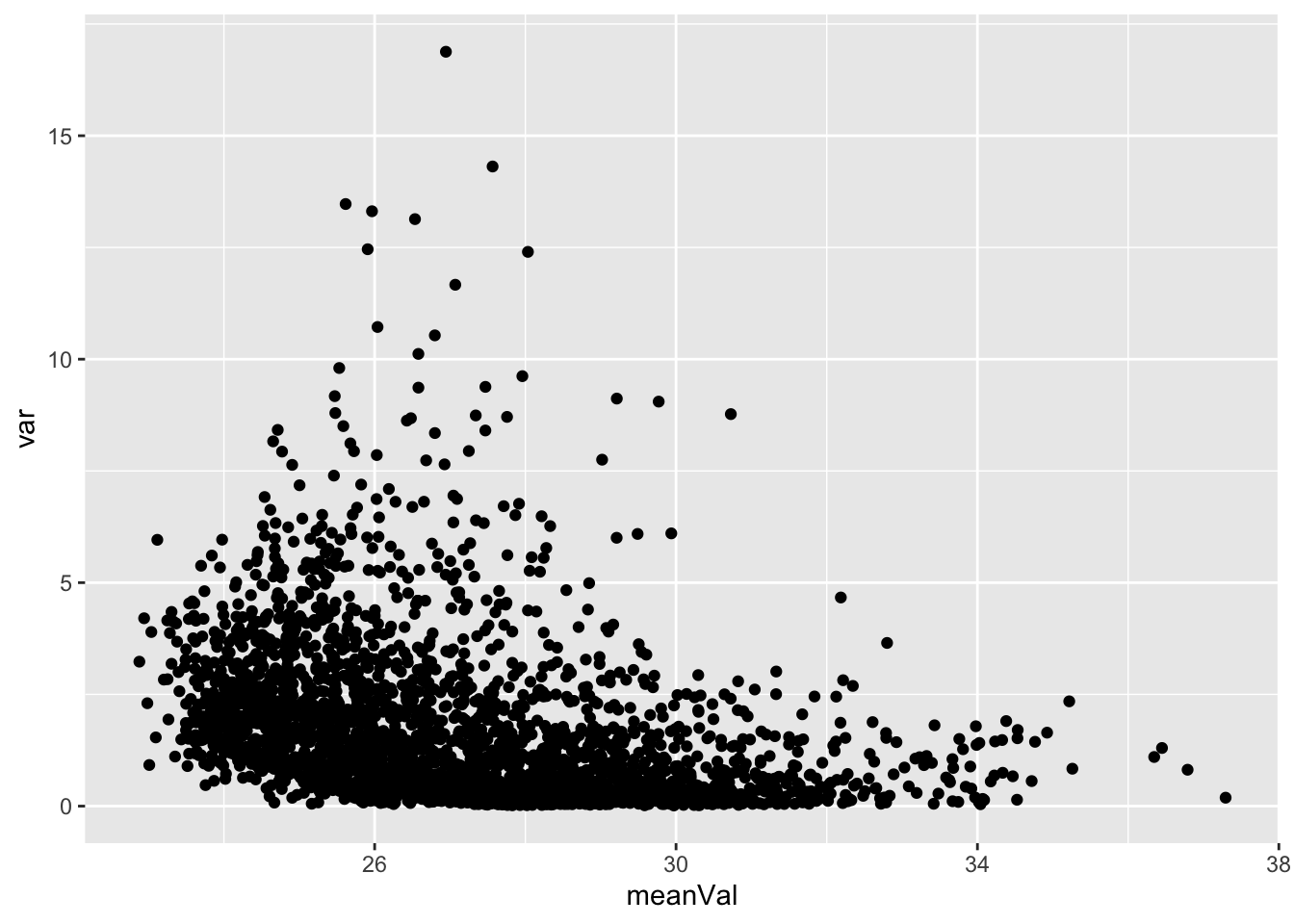
Heatmap visualization
library(pheatmap)
#select top 1000 most variant
colAnno <- colData(protFilt) %>% data.frame()
#colAnno[["sampleName"]] <- NULL
plotMat <- countMat[order(plotTab$var, decreasing = TRUE)[1:1000],]
pheatmap(plotMat, show_rownames = FALSE, scale = "row",
annotation_col = colAnno,
clustering_method = "ward.D2")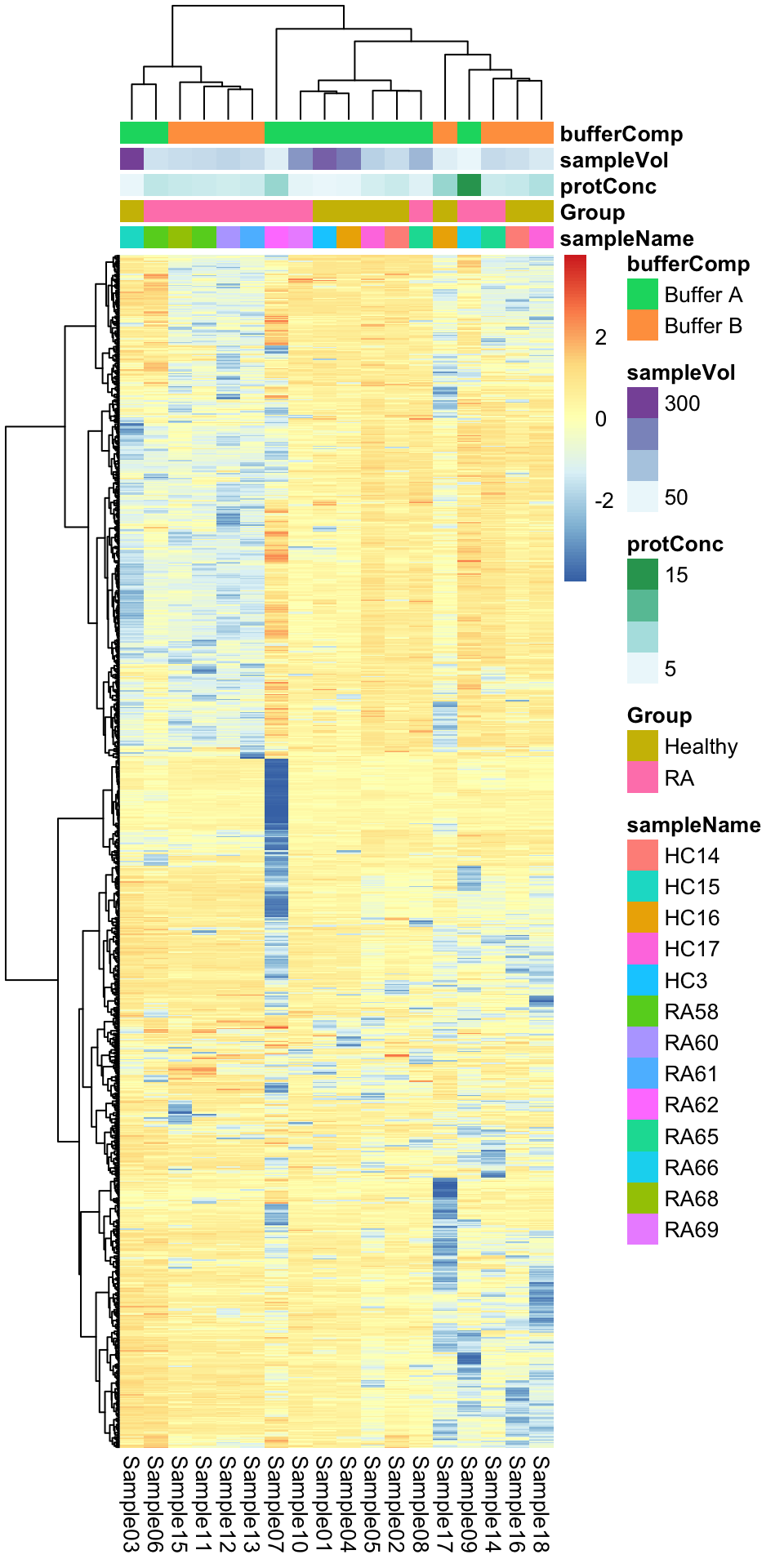
PCA
prRes <- prcomp(t(plotMat), scale. = TRUE, center = TRUE)
plotTab <- prRes$x %>% as_tibble(rownames = "sampleID") %>%
left_join(as_tibble(colAnno, rownames = "sampleID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = Group, shape = bufferComp)) +
geom_point() +
ggrepel::geom_text_repel(aes(label = sampleName))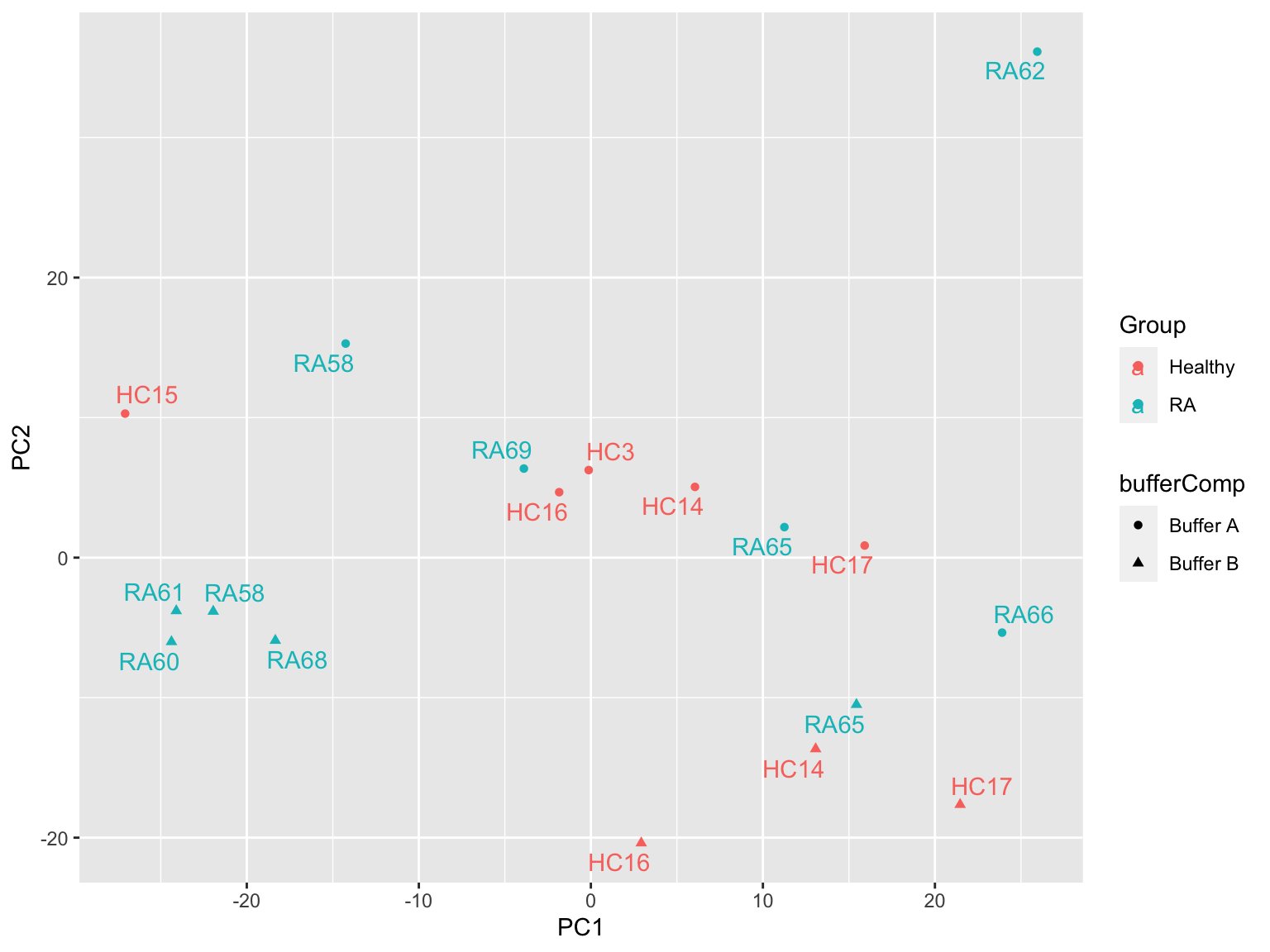 Buffer composition may act as a confounding factor. One sample, RA62 may
be outlier.
Buffer composition may act as a confounding factor. One sample, RA62 may
be outlier.
Deal with technical replicates
Based on PCA, if there’s a replicate, choose buffer A
smpTab <- colData(protFilt) %>% as_tibble(rownames = "sampleID") %>%
arrange(bufferComp, sampleName) %>% distinct(sampleName, .keep_all = TRUE)
protSub <- protFilt[,smpTab$sampleID]Remove one potential outlier, RA62
protSub <- protSub[,protSub$sampleName != "RA62"]Differential expression using proDA
protSub$Group <- factor(protSub$Group)
table(protSub$Group)
Healthy RA
5 7 Only keep proteins with symbols
protSub <- protSub[!rowData(protSub)$name %in% c("",NA),]Differential protein expression using proDA
library(proDA)
protMat <- assays(protSub)[["norm"]]
fit <- proDA(protMat, design = ~ Group,
col_data = colData(protSub))
resTab <- test_diff(fit, contrast = "GroupRA") %>%
arrange(pval) %>%
mutate(symbol = rowData(protSub[name,])$name)hist(resTab$pval)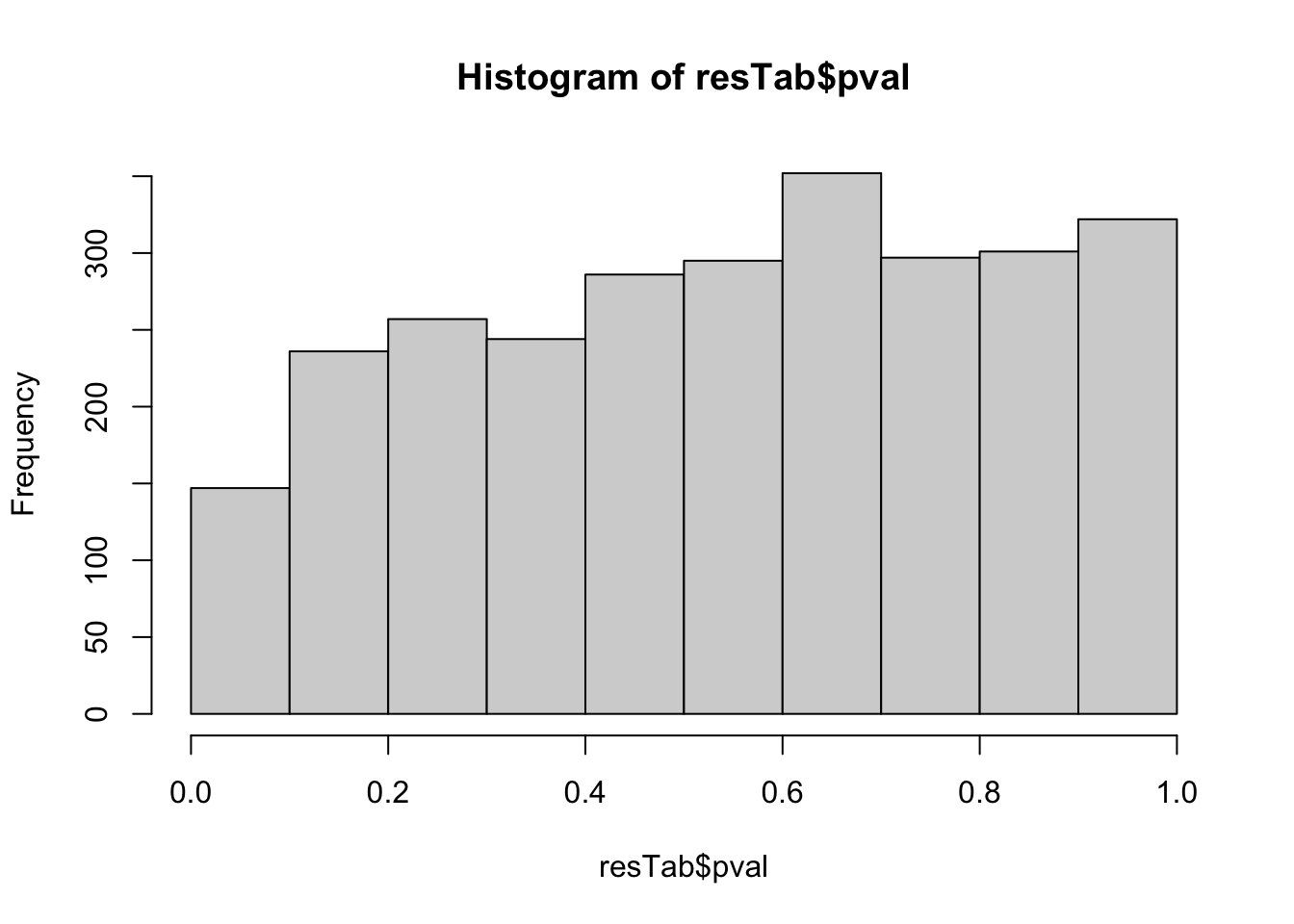 Not strong difference
Not strong difference
Proteins with p-value < 0.05
resTab.sig <- filter(resTab, pval < 0.05)
resTab.sig %>% select(symbol, pval, adj_pval, diff) %>%
mutate_if(is.numeric, formatC, digits=1) %>%
DT::datatable()Plot top 9 examples
pList <- lapply(seq(9), function(i) {
rec <- resTab.sig[i,]
plotTab <- tibble(expr = protMat[rec$name,],
Group = protSub$Group)
ggplot(plotTab, aes(x=Group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = Group), size=3) +
ggtitle(rec$symbol) +
theme_bw()
})
cowplot::plot_grid(plotlist = pList,ncol=3)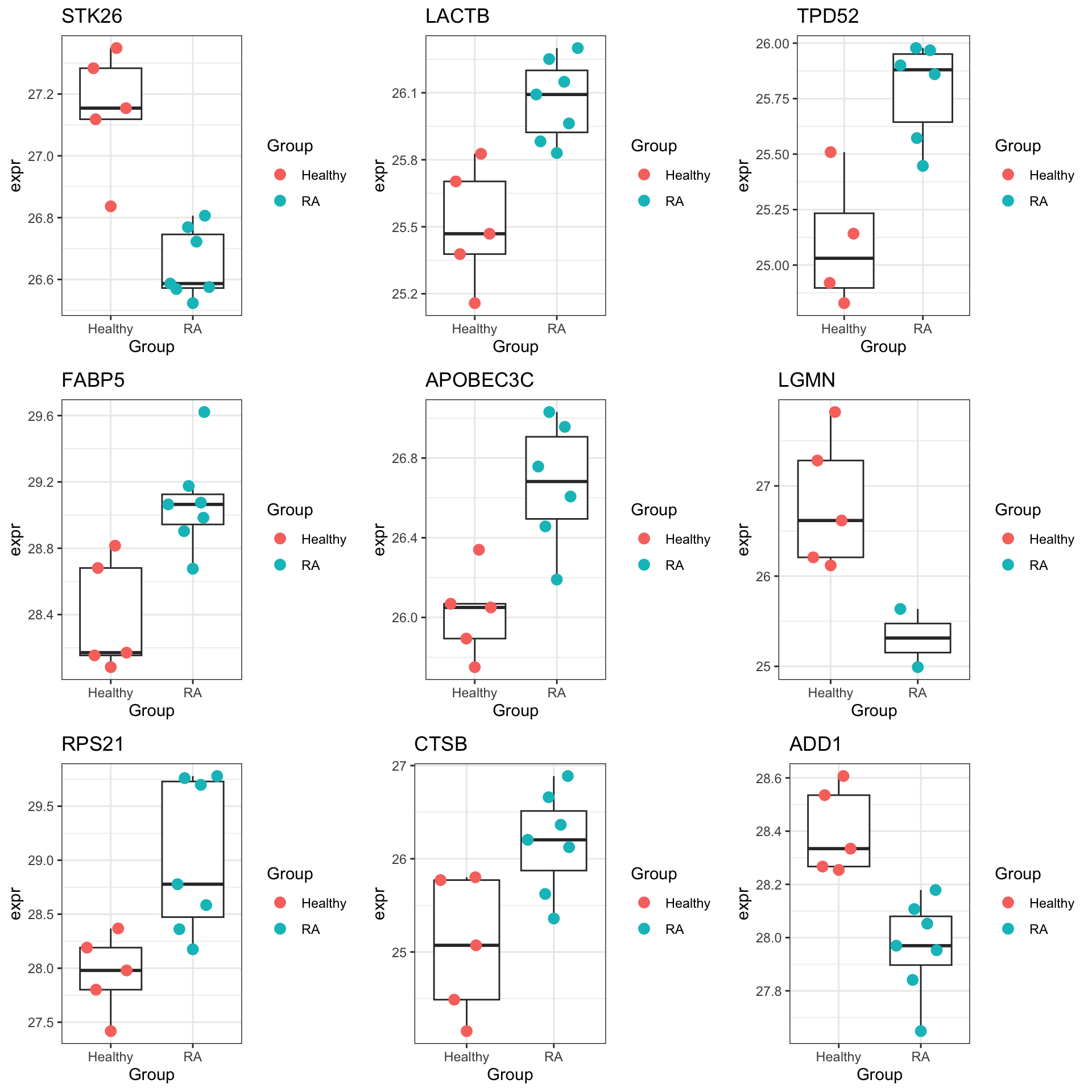
Enrichment analysis
gmts = list(H= "~/CLLproject_jlu/data/commonFiles/h.all.v6.2.symbols.gmt",
KEGG = "~/CLLproject_jlu/data/commonFiles/c2.cp.kegg.v6.2.symbols.gmt",
C6 = "~/CLLproject_jlu/data/commonFiles/c6.all.v6.2.symbols.gmt")
inputTab <- resTab %>% filter(pval < 0.1) %>%
distinct(symbol, .keep_all = TRUE) %>%
select(symbol, t_statistic) %>% data.frame() %>% column_to_rownames("symbol")
enRes <- list()
enRes[["Hallmark"]] <- runGSEA(inputTab, gmts$H, "page")
enRes[["KEGG"]] <- runGSEA(inputTab, gmts$KEGG,"page")
enRes[["Perturbation"]] <- runGSEA(inputTab, gmts$C6,"page")
p <- plotEnrichmentBar(enRes, pCut =0.05, ifFDR= FALSE)
cowplot::plot_grid(p)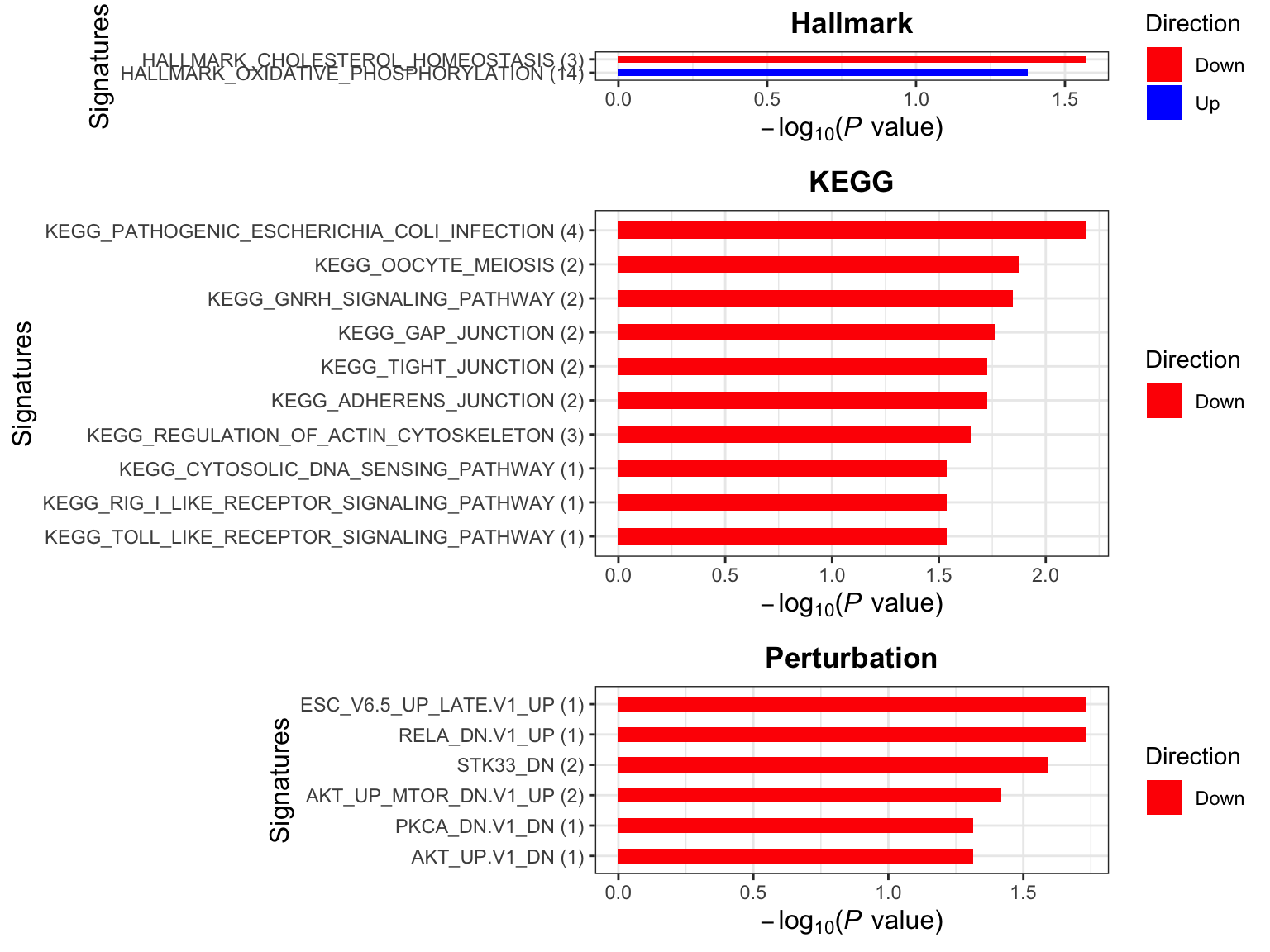
Plot expression of proteins from OXPHOS pathway
geneList <- piano::loadGSC(gmts$H)$gsc
resTab.oxphos <- filter(resTab, pval < 0.1,
symbol %in% geneList$HALLMARK_OXIDATIVE_PHOSPHORYLATION)
pList <- lapply(seq(nrow(resTab.oxphos)), function(i) {
rec <- resTab.oxphos[i,]
plotTab <- tibble(expr = protMat[rec$name,],
Group = protSub$Group)
ggplot(plotTab, aes(x=Group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = Group), size=3) +
ggtitle(rec$symbol) +
theme_bw()
})
cowplot::plot_grid(plotlist = pList,ncol=3)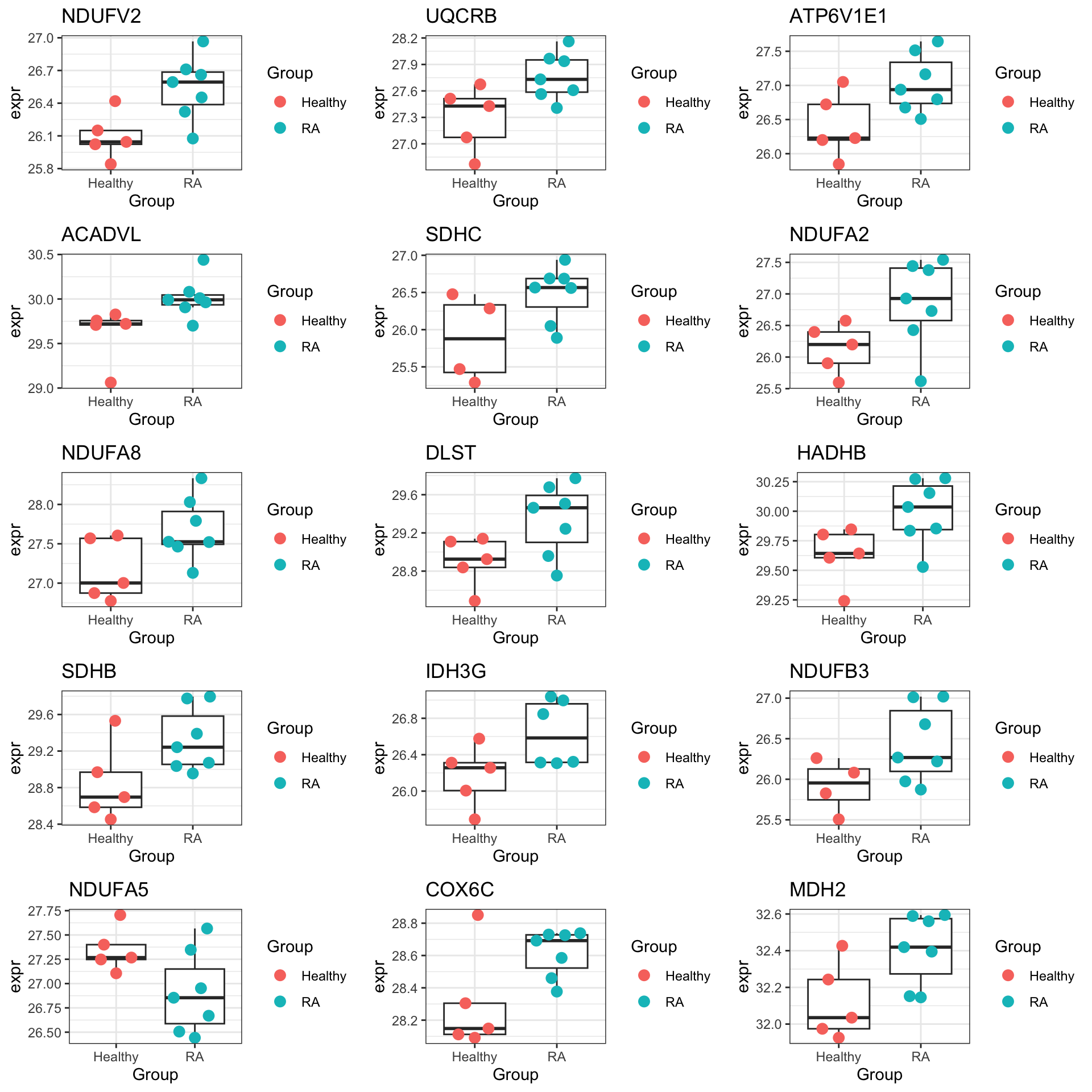
DNMT1 expression
countMat <- assays(protSub)[["imputed"]]
resDNMT1 <- filter(resTab, str_detect(symbol,"DNMT1"))
plotTab <- tibble(expr = countMat[resDNMT1$name,],
Group = protSub$Group)
ggplot(plotTab, aes(x=Group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = Group), size=3) +
ggtitle("DNMT1") +
theme_bw()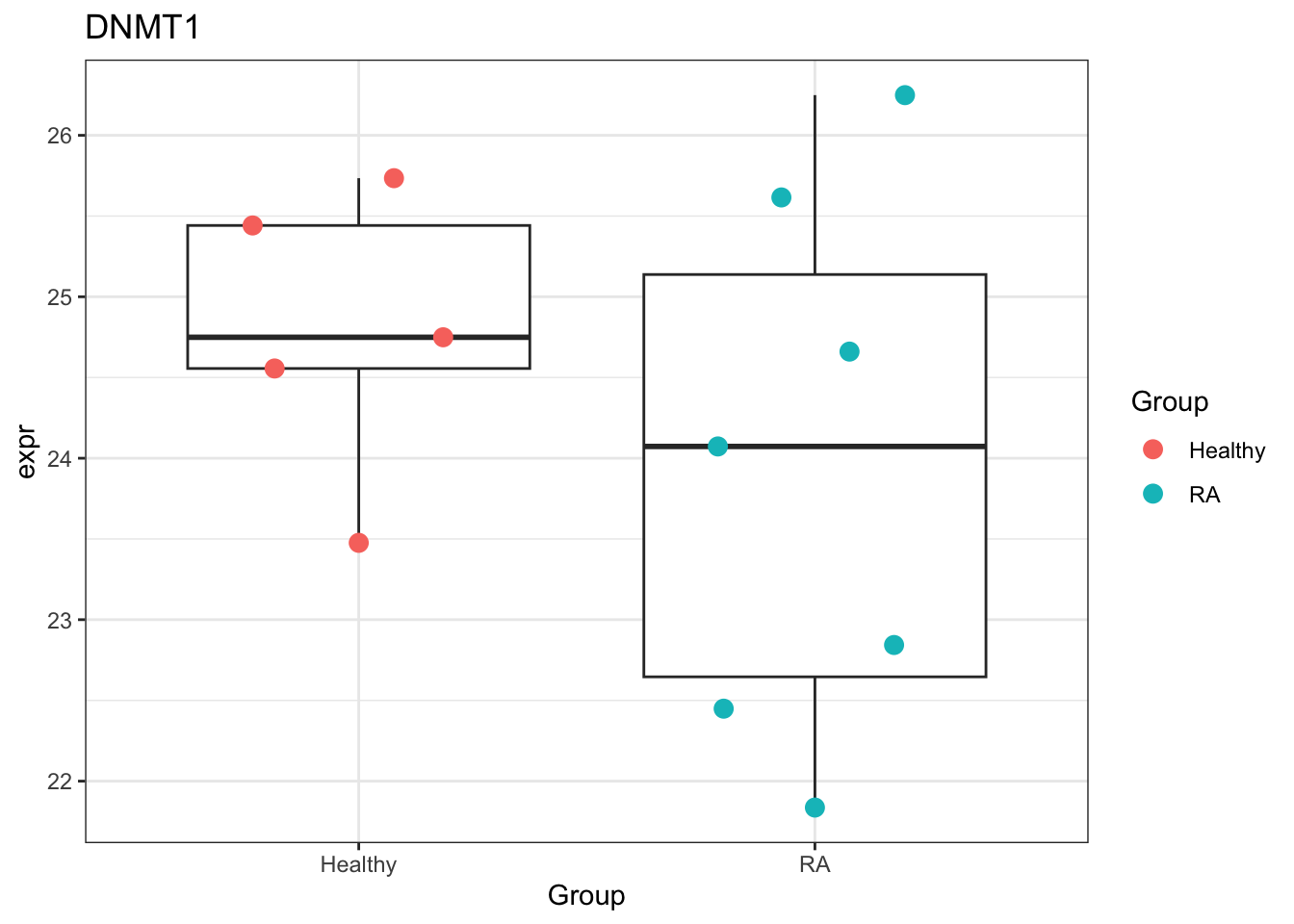
Power analysis to estimate sample size
rowData(protSub)$imputed <- NULL
protTab <- jyluMisc::sumToTidy(protSub)
meanTab <- protTab %>%
group_by(Group, ID) %>%
summarise(mean = mean(imputed)) %>%
pivot_wider(names_from = Group, values_from = mean)
sdTab <- protTab %>%
group_by(ID) %>%
summarise(sd = sd(imputed))
dTab <- left_join(meanTab, sdTab) %>%
mutate(d= abs(RA-Healthy)/sd)nTab <- lapply(seq(nrow(dTab)), function(i) {
rec <- dTab[i,]
n1 <- pwr.t.test(d = rec$d , sig.level = 0.05,
power = 0.9, alternative = "two.sided")$n
n2 <- pwr.t.test(d = rec$d , sig.level = 0.01,
power = 0.9, alternative = "two.sided")$n
tibble(ID = rec$ID,
n0.05 = as.integer(n1), n0.01=as.integer(n2))
}) %>% bind_rows() %>%
arrange(n0.05) %>%
mutate(name = rowData(protSub[ID,])$name)Sample size table
nTab %>% select(-ID) %>%
DT::datatable()Sample size versus DE proteins detectable.
plotTab <- lapply(seq(0.01, 1.5, length.out=100), function(x) {
tibble(nCall = nrow(filter(dTab, d > x)),
nSample = pwr.t.test(d = x , sig.level = 0.05,
power = 0.8, type = "two.sample")$n)
}) %>% bind_rows() %>%
arrange(nCall) %>% filter(nCall >0)
ggplot(plotTab, aes(x=nCall, y=nSample)) +
geom_line() + geom_point() +
ylim(0,100) + xlim(0,1500) +
ylab("sample per group") +
xlab("differentially expressed proteins at P=0.05") +
theme_bw() 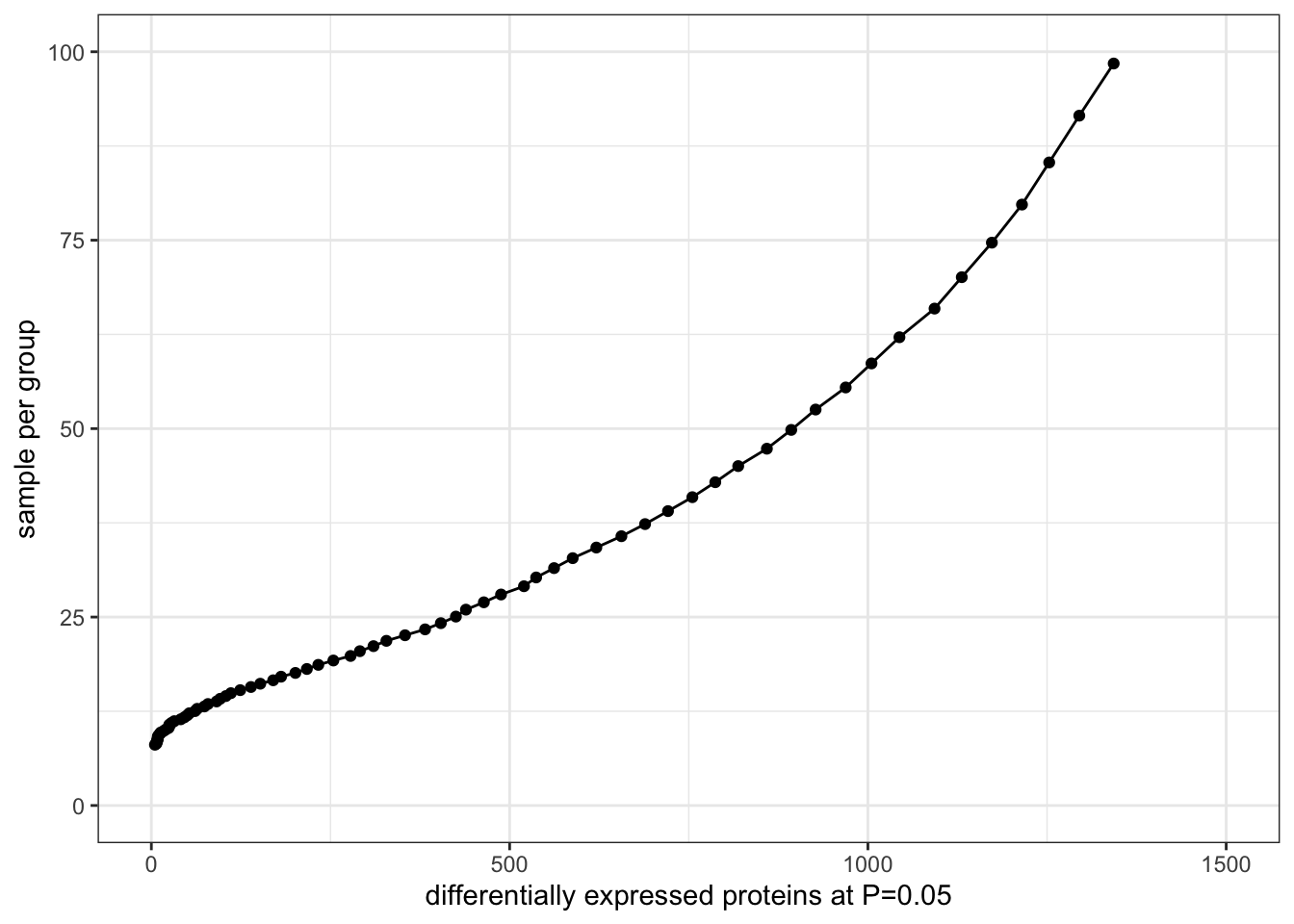 (n = sample in each group)
(n = sample in each group)
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] piano_2.12.0 proDA_1.10.0
[3] pheatmap_1.0.12 limma_3.52.2
[5] forcats_0.5.1 stringr_1.4.1
[7] dplyr_1.0.9 purrr_0.3.4
[9] readr_2.1.2 tidyr_1.2.0
[11] tibble_3.1.8 ggplot2_3.4.1
[13] tidyverse_1.3.2 SummarizedExperiment_1.26.1
[15] GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[17] IRanges_2.30.0 S4Vectors_0.34.0
[19] MatrixGenerics_1.8.1 matrixStats_0.62.0
[21] jyluMisc_0.1.5 pwr_1.3-0
[23] vsn_3.64.0 Biobase_2.56.0
[25] BiocGenerics_0.42.0
loaded via a namespace (and not attached):
[1] DEP_1.18.0 utf8_1.2.2 shinydashboard_0.7.2
[4] gmm_1.6-6 tidyselect_1.1.2 htmlwidgets_1.5.4
[7] grid_4.2.0 BiocParallel_1.30.3 norm_1.0-10.0
[10] maxstat_0.7-25 munsell_0.5.0 codetools_0.2-18
[13] preprocessCore_1.58.0 DT_0.23 withr_2.5.0
[16] colorspace_2.0-3 highr_0.9 knitr_1.39
[19] rstudioapi_0.13 ggsignif_0.6.3 mzID_1.34.0
[22] labeling_0.4.2 git2r_0.30.1 slam_0.1-50
[25] GenomeInfoDbData_1.2.8 KMsurv_0.1-5 farver_2.1.1
[28] rprojroot_2.0.3 vctrs_0.5.2 generics_0.1.3
[31] TH.data_1.1-1 xfun_0.31 sets_1.0-21
[34] R6_2.5.1 doParallel_1.0.17 ggbeeswarm_0.6.0
[37] clue_0.3-61 MsCoreUtils_1.8.0 bitops_1.0-7
[40] cachem_1.0.6 fgsea_1.22.0 DelayedArray_0.22.0
[43] assertthat_0.2.1 promises_1.2.0.1 scales_1.2.0
[46] multcomp_1.4-19 googlesheets4_1.0.0 beeswarm_0.4.0
[49] gtable_0.3.0 extraDistr_1.9.1 Cairo_1.6-0
[52] affy_1.74.0 sandwich_3.0-2 workflowr_1.7.0
[55] rlang_1.0.6 mzR_2.30.0 GlobalOptions_0.1.2
[58] splines_4.2.0 rstatix_0.7.0 impute_1.70.0
[61] gargle_1.2.0 broom_1.0.0 BiocManager_1.30.18
[64] yaml_2.3.5 abind_1.4-5 modelr_0.1.8
[67] crosstalk_1.2.0 backports_1.4.1 httpuv_1.6.6
[70] tools_4.2.0 relations_0.6-12 affyio_1.66.0
[73] ellipsis_0.3.2 gplots_3.1.3 jquerylib_0.1.4
[76] RColorBrewer_1.1-3 MSnbase_2.22.0 plyr_1.8.7
[79] Rcpp_1.0.9 visNetwork_2.1.0 zlibbioc_1.42.0
[82] RCurl_1.98-1.7 ggpubr_0.4.0 GetoptLong_1.0.5
[85] cowplot_1.1.1 zoo_1.8-10 ggrepel_0.9.1
[88] haven_2.5.0 cluster_2.1.3 exactRankTests_0.8-35
[91] fs_1.5.2 magrittr_2.0.3 magick_2.7.3
[94] data.table_1.14.2 circlize_0.4.15 reprex_2.0.1
[97] survminer_0.4.9 pcaMethods_1.88.0 googledrive_2.0.0
[100] mvtnorm_1.1-3 ProtGenerics_1.28.0 hms_1.1.1
[103] shinyjs_2.1.0 mime_0.12 evaluate_0.15
[106] xtable_1.8-4 XML_3.99-0.10 readxl_1.4.0
[109] gridExtra_2.3 shape_1.4.6 compiler_4.2.0
[112] ncdf4_1.19 KernSmooth_2.23-20 crayon_1.5.2
[115] htmltools_0.5.4 later_1.3.0 tzdb_0.3.0
[118] lubridate_1.8.0 DBI_1.1.3 dbplyr_2.2.1
[121] ComplexHeatmap_2.12.0 tmvtnorm_1.5 MASS_7.3-58
[124] Matrix_1.4-1 car_3.1-0 cli_3.4.1
[127] imputeLCMD_2.1 marray_1.74.0 parallel_4.2.0
[130] igraph_1.3.4 pkgconfig_2.0.3 km.ci_0.5-6
[133] MALDIquant_1.21 xml2_1.3.3 foreach_1.5.2
[136] vipor_0.4.5 bslib_0.4.1 XVector_0.36.0
[139] drc_3.0-1 rvest_1.0.2 digest_0.6.30
[142] rmarkdown_2.14 cellranger_1.1.0 fastmatch_1.1-3
[145] survMisc_0.5.6 shiny_1.7.4 gtools_3.9.3
[148] rjson_0.2.21 lifecycle_1.0.3 jsonlite_1.8.3
[151] carData_3.0-5 fansi_1.0.3 pillar_1.8.0
[154] lattice_0.20-45 fastmap_1.1.0 httr_1.4.3
[157] plotrix_3.8-2 survival_3.4-0 glue_1.6.2
[160] png_0.1-7 iterators_1.0.14 stringi_1.7.8
[163] sass_0.4.2 caTools_1.18.2