Explore and analyze each individual single omic dataset
Junyan Lu
Last updated: 2024-11-25
Checks: 4 2
Knit directory: RA_Tcell_omics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221110) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- unnamed-chunk-10
- unnamed-chunk-14
- unnamed-chunk-15
- unnamed-chunk-21
- unnamed-chunk-22
- unnamed-chunk-32
- unnamed-chunk-9
To ensure reproducibility of the results, delete the cache directory
singleOmic_analysis_cache and re-run the analysis. To have
workflowr automatically delete the cache directory prior to building the
file, set delete_cache = TRUE when running
wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the
results is critical for reproducibility. To start using Git, open the
Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full
reproducibility benefits of using workflowr, please see
?wflow_start.
Load libraries
Load processed data
load("../output/maeObj.RData")Analysis of metabolomic data
seMeta <- maeObj[["Metabolism"]]
metaMat <- assay(seMeta)
metaTab <- sumToTidy(seMeta)Features and numbers
dim(seMeta)[1] 65 36Visualize overall data distribution
Per sample
ggplot(metaTab, aes(x=colID, y=glog(count))) +
geom_boxplot() + geom_point(aes(col = group)) +
theme(axis.text = element_text(angle = 90, hjust = 1, vjust = 0.5))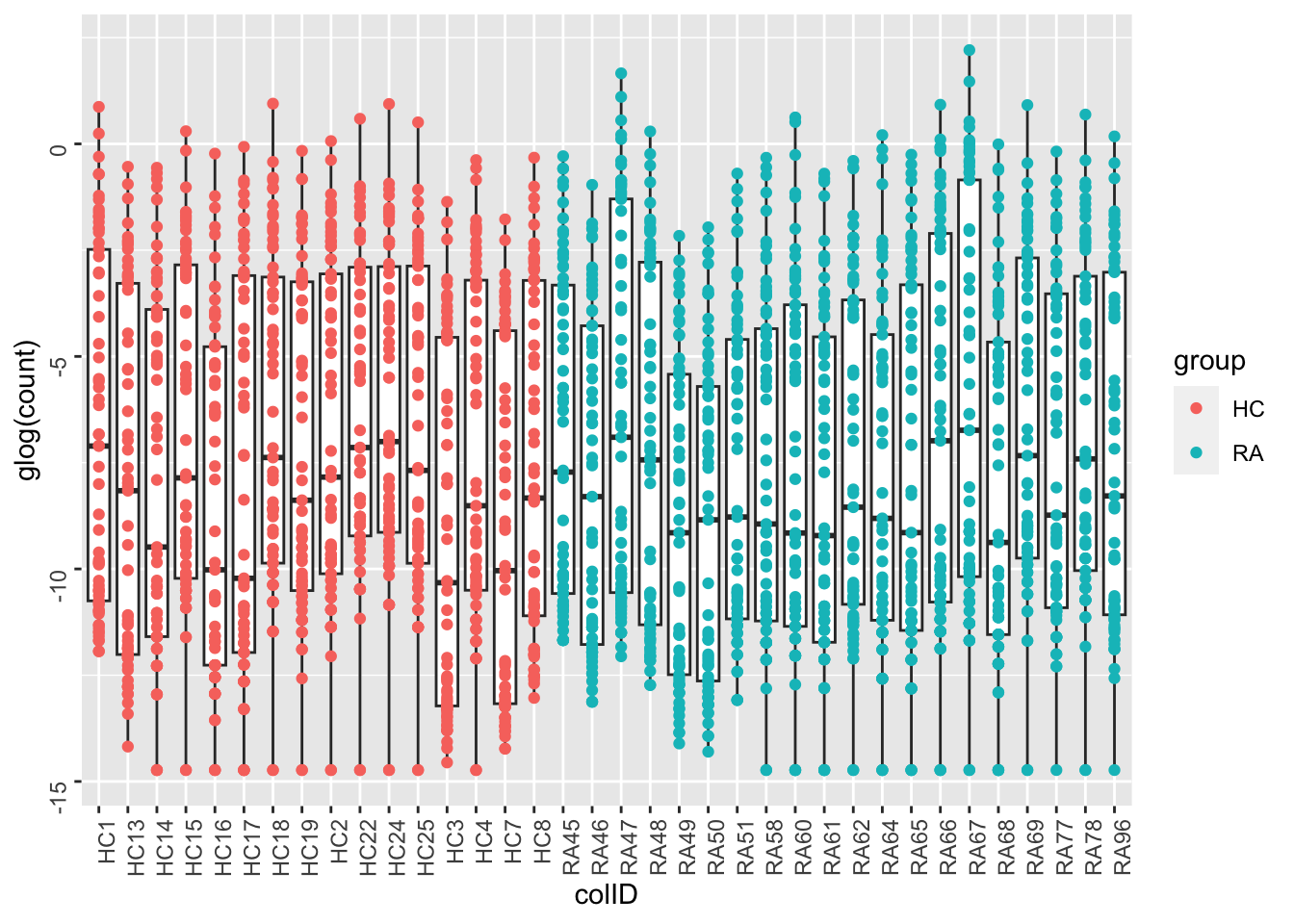
Color by experiment date
ggplot(metaTab, aes(x=colID, y=glog(count))) +
geom_boxplot() + geom_point(aes(col = dateMeta)) +
theme(axis.text = element_text(angle = 90, hjust = 1, vjust = 0.5))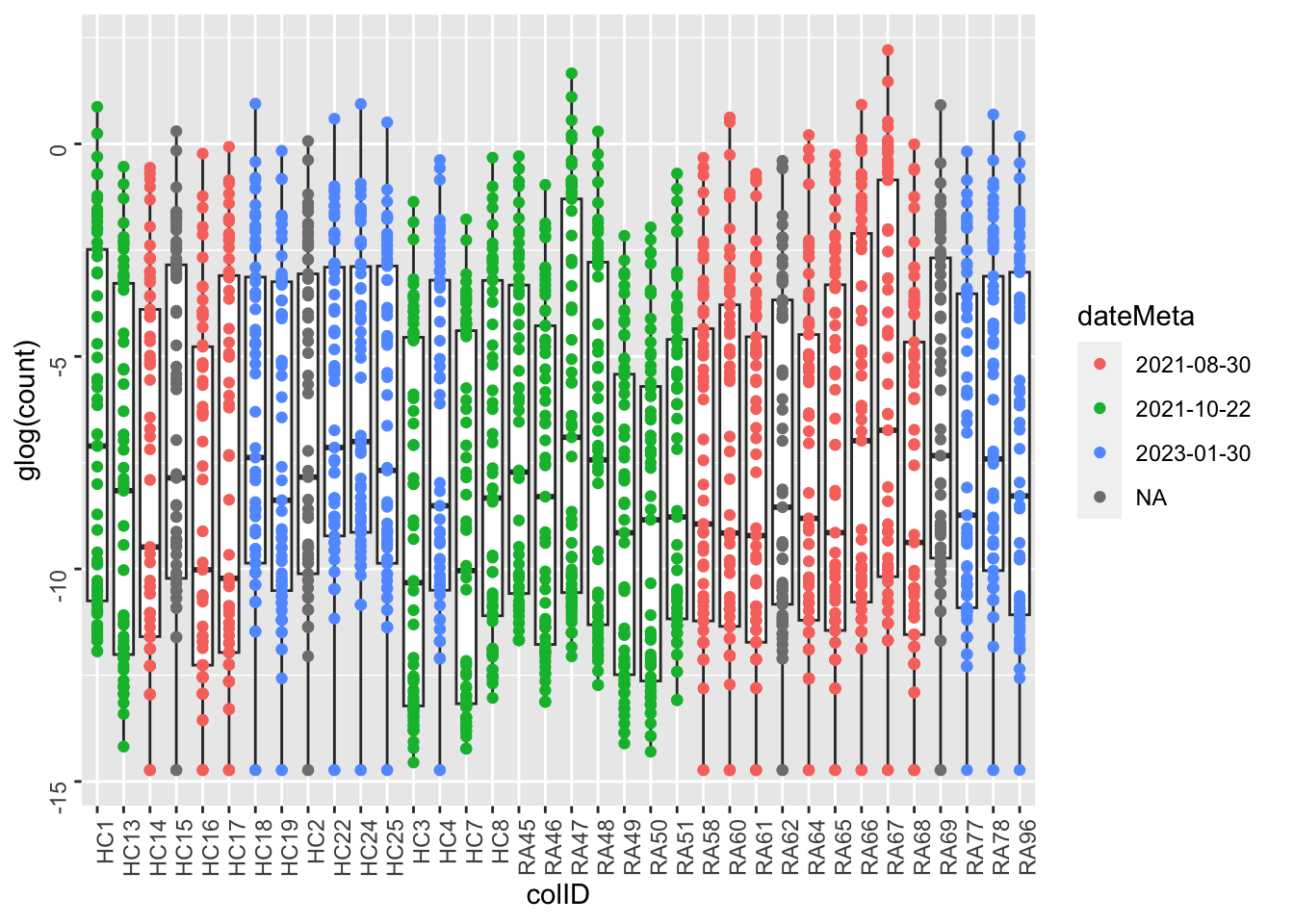
Per metabolite
ggplot(metaTab, aes(x=metabolite, y=glog(count))) +
geom_boxplot() + geom_point(aes(col = group)) +
theme(axis.text = element_text(angle = 90, hjust = 1, vjust = 0.5))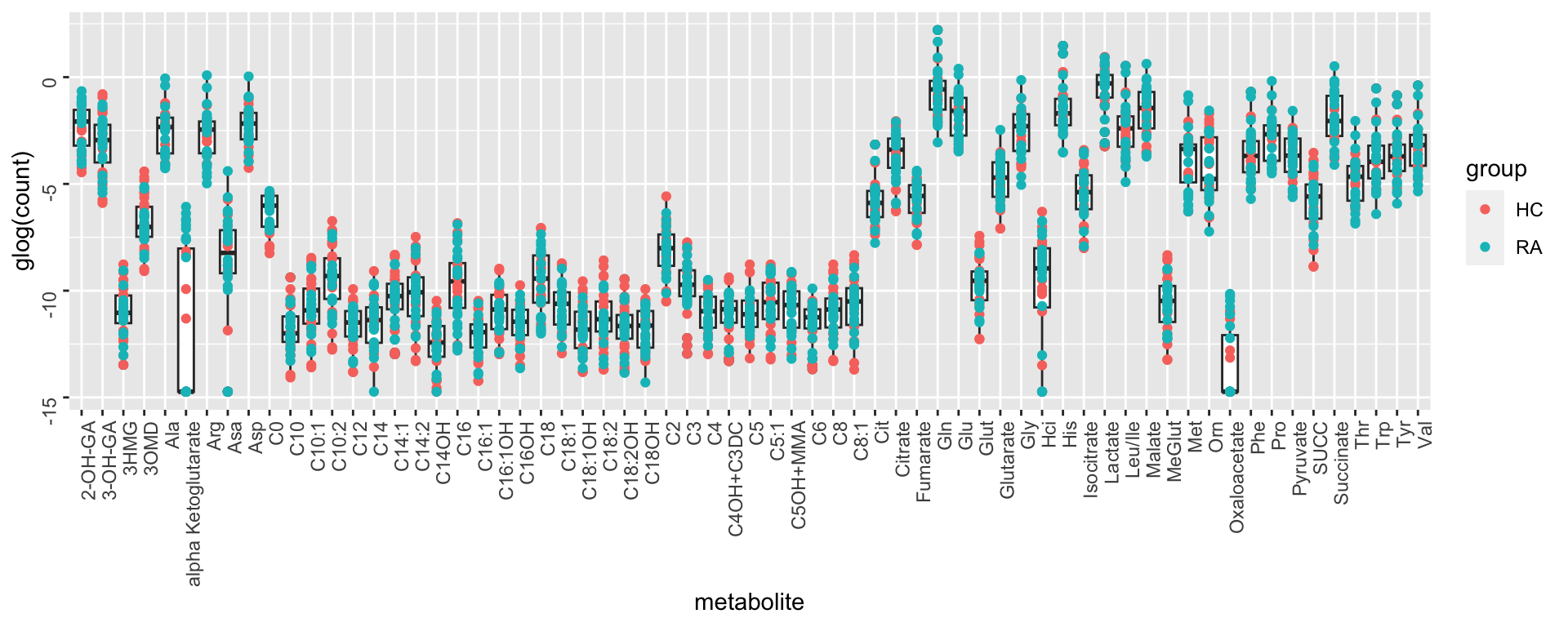 The abundance of different metabolites are very different.
Transformation and Normalization may not be needed actually
The abundance of different metabolites are very different.
Transformation and Normalization may not be needed actually
PCA
By group
metaMat.scale <- glog(metaMat)
pcRes <- prcomp(t(metaMat.scale), scale. = TRUE, center = TRUE)$x
plotTab <- as_tibble(pcRes, rownames = "colID") %>%
left_join(as.tibble(colData(seMeta), rownames = "colID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = group)) +
geom_point()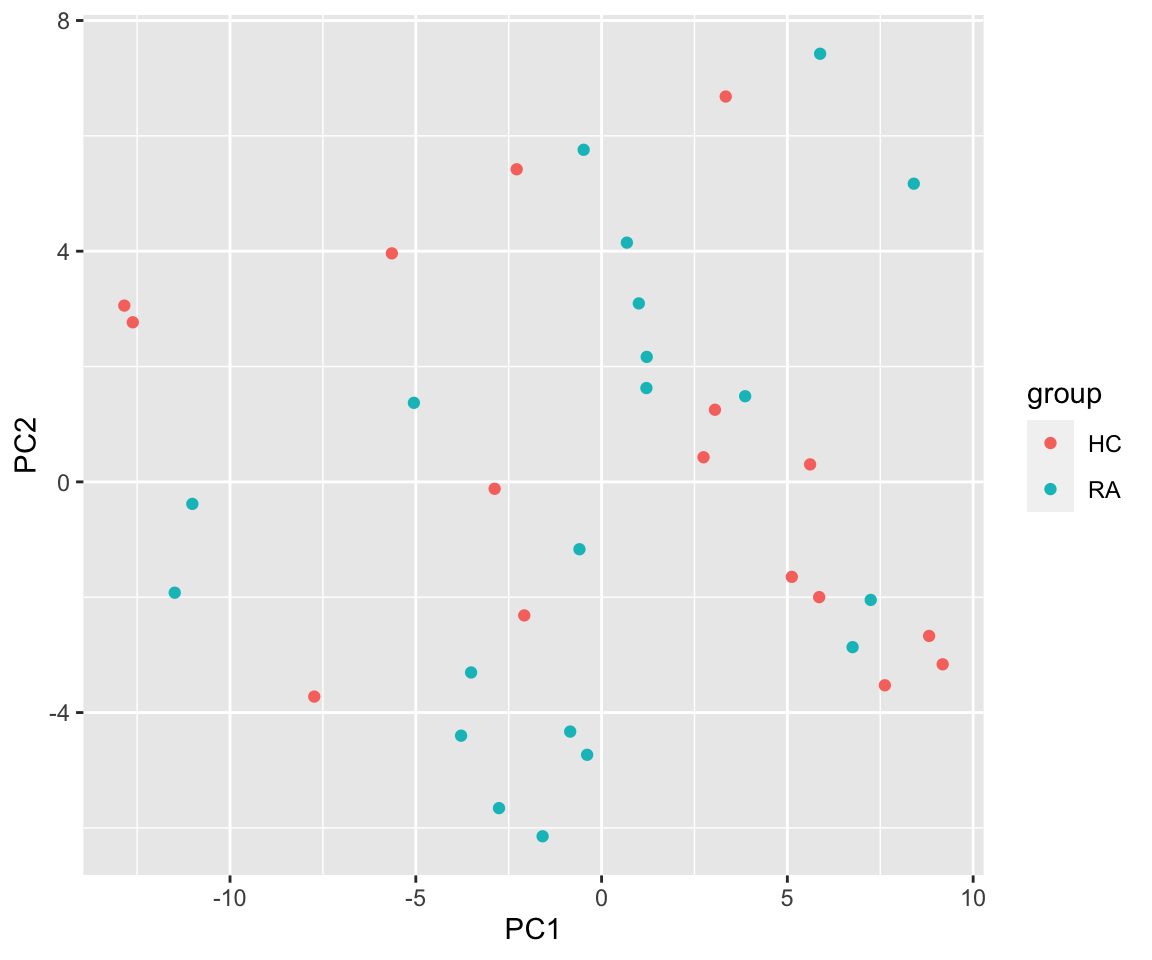
By date
metaMat.scale <- glog(metaMat)
pcRes <- prcomp(t(metaMat.scale), scale. = TRUE, center = TRUE)$x
plotTab <- as_tibble(pcRes, rownames = "colID") %>%
left_join(as.tibble(colData(seMeta), rownames = "colID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = dateMeta)) +
geom_point()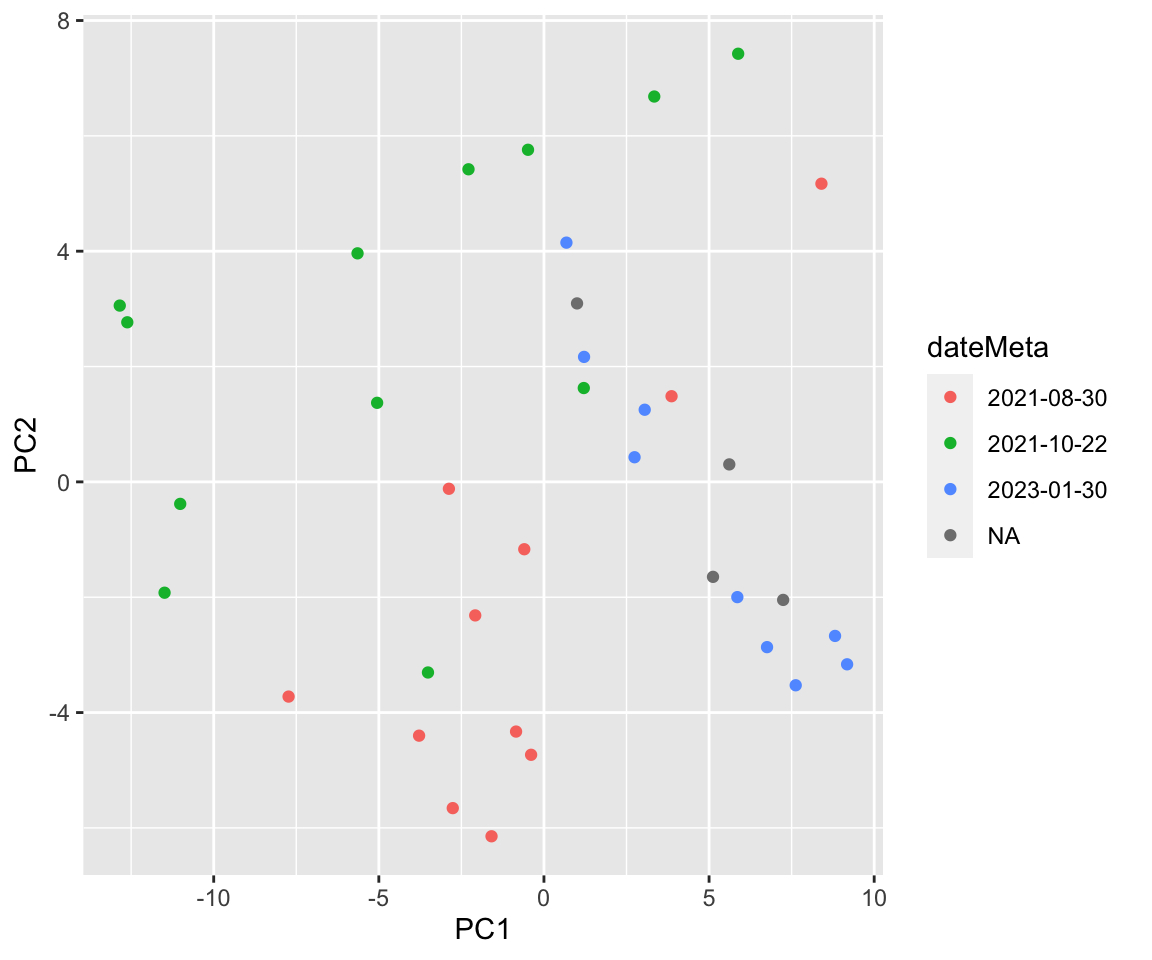 There are potentially some batch effect in the metabolic
dataset
There are potentially some batch effect in the metabolic
dataset
table(seMeta$group, seMeta$dateMeta)
2021-08-30 2021-10-22 2023-01-30
HC 3 5 6
RA 8 7 3Differential abundance test
With correction for batch effect. Two samples do not have date information
seMetaSub <- seMeta[,!is.na(seMeta$dateMeta)]
#assay(seMetaSub) <- glog(assay(seMetaSub))
assays(seMetaSub)[["combat"]] <- sva::ComBat(assay(seMetaSub), batch = seMetaSub$dateMeta)Found 2 genes with uniform expression within a single batch (all zeros); these will not be adjusted for batch.library(limma)
metaMat <- assays(seMetaSub)[["combat"]]
designMat <- model.matrix(~group, data = colData(seMetaSub))
lmFit <- lmFit(metaMat, design = designMat)
fit2 <- eBayes(lmFit)
resTab <- topTable(fit2, number = Inf) %>%
as_tibble(rownames = "metabolite")
hist(resTab$P.Value)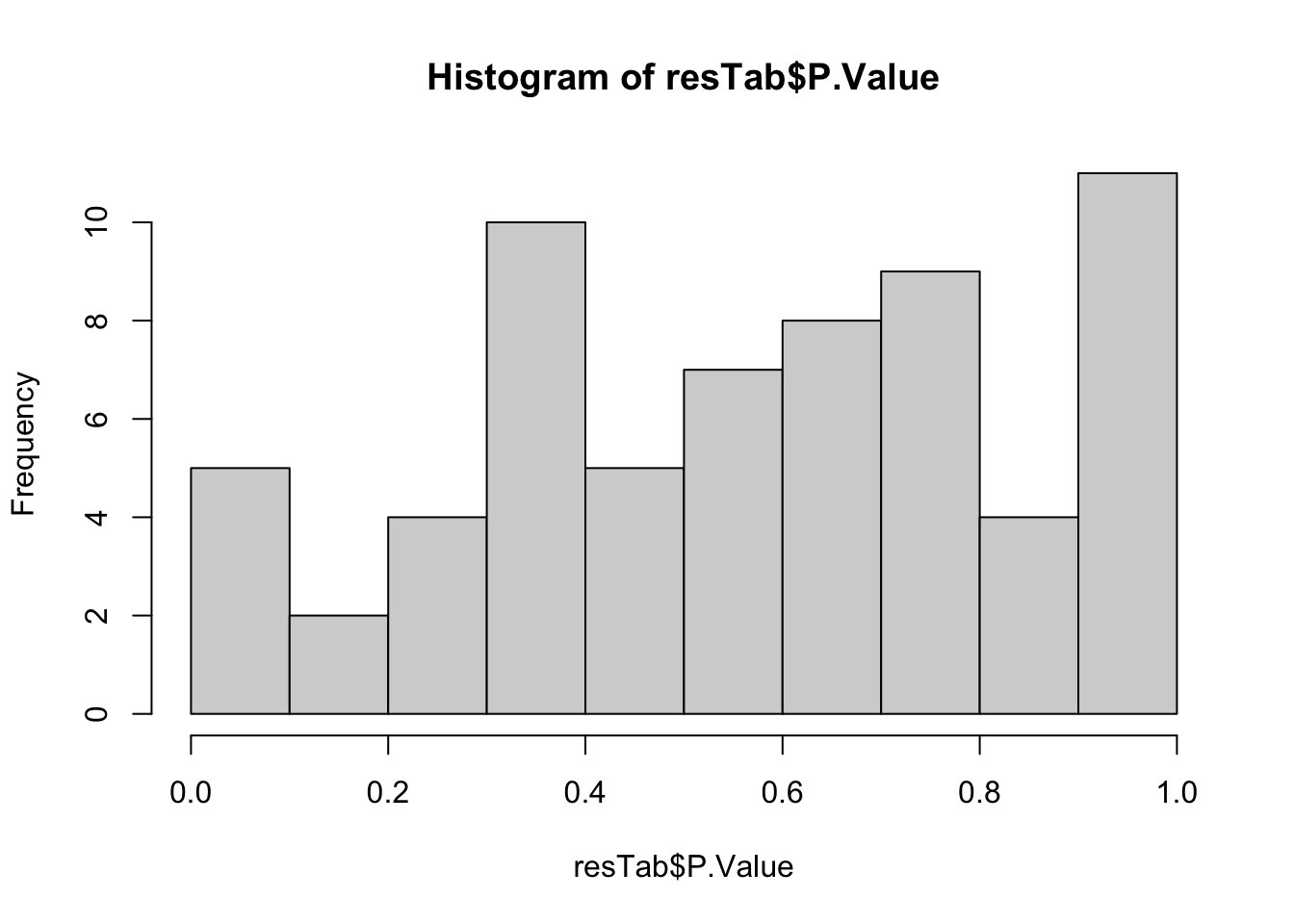
Plot significant associations
pList <- lapply(seq(nrow(filter(resTab, P.Value <= 0.1))), function(i) {
rec <- resTab[i,]
plotTab <- filter(metaTab, metabolite == rec$metabolite) %>%
mutate(group=ifelse(group == "HC","Control",group))
#plotTab <- tibble(colID = colnames(metaMat),
# count = metaMat[rec$metabolite,]) %>%
# mutate(group = seMeta[,colID]$group)
ggplot(plotTab, aes(x=group, y=count)) +
geom_boxplot(outlier.shape = NA, width =0.3) +
ggbeeswarm::geom_quasirandom(aes(color = group), size=3, width = 0.3) +
ggtitle(sprintf("%s\n(P=%s)",rec$metabolite,formatC(rec$P.Value,digits = 2))) +
scale_color_manual(values =c(Control = "blue", RA = "red")) +
theme_classic() +
theme(legend.position = "none",
axis.text = element_text(face = "bold", size=14), axis.title = element_text(face = "bold",size=14),
plot.title = element_text(hjust=.5, face = "bold")) +
ylab("log (abundance)") + xlab("")
})
cowplot::plot_grid(plotlist = pList,ncol=3)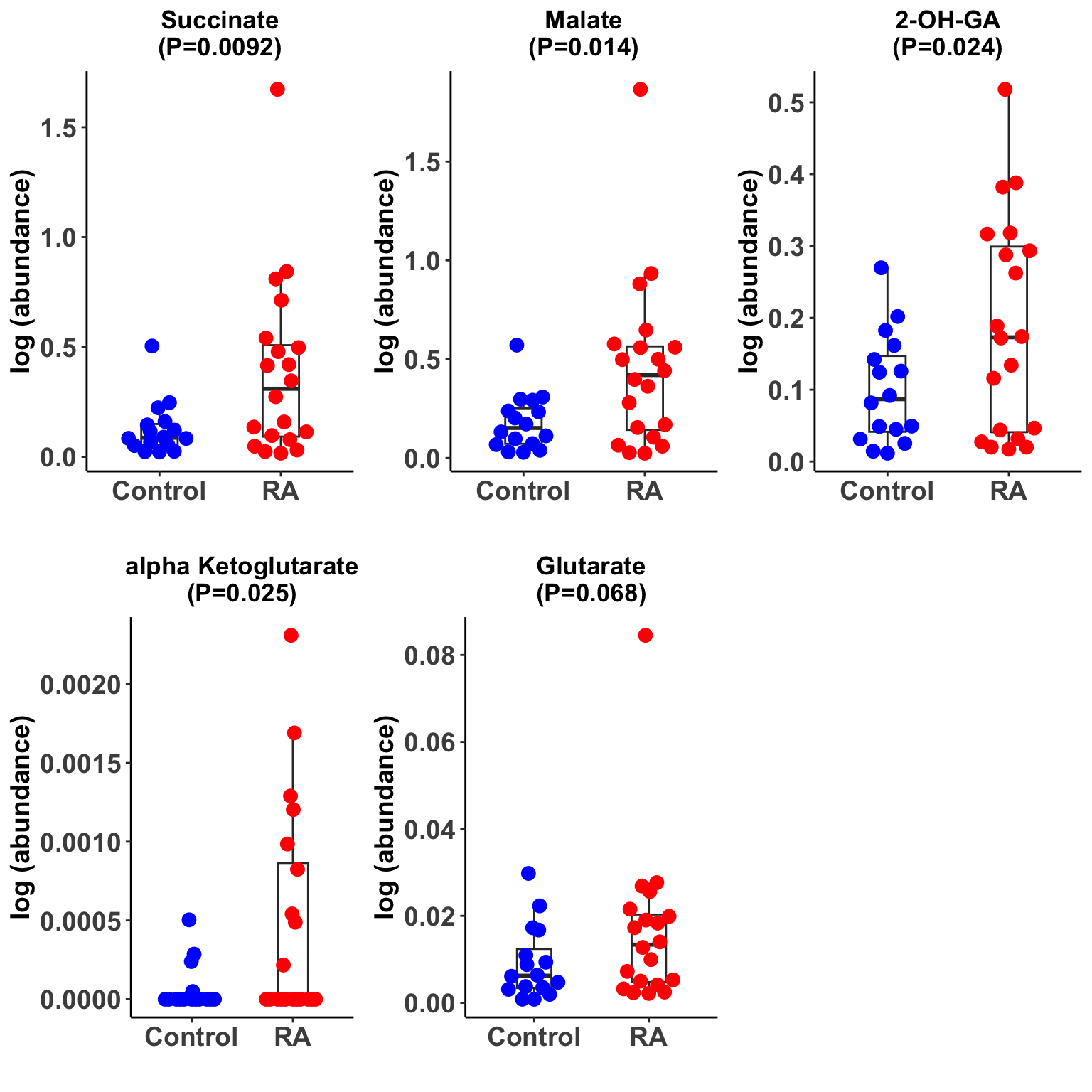
ggsave("../docs/meta_sig_boxplot.pdf", height = 8, width = 8)Heatmap visualization
All features
library(pheatmap)
#select top 1000 most variant
colAnno <- colData(seMeta)[,"group",drop=FALSE] %>%
data.frame()
colAnno$group <- str_replace(colAnno$group, "HC","Control")
seMetaSub <- seMeta[,!is.na(seMeta$dateMeta)]
assay(seMetaSub) <- glog(assay(seMetaSub))
assays(seMetaSub)[["combat"]] <- sva::ComBat(assay(seMetaSub), batch = seMetaSub$dateMeta)
plotMat <- assays(seMetaSub)[["combat"]]
annoColor <- list(group = c(Control = "blue", RA = "red"))
pdf("../docs/meta_heatmap_all.pdf",height = 10, width = 10)
pheatmap(plotMat, scale = "row",
annotation_col = colAnno,
cluster_rows = T, cluster_cols = FALSE,
color = colorRampPalette(c("navy", "white", "firebrick"))(100),
breaks = seq(-3,3,length.out=100),
annotation_colors = annoColor, annotation_names_col = FALSE, fontsize_row = 11)
dev.off()Only features with p-value < 0.25
library(pheatmap)
#select top 1000 most variant
#colAnno <- colData(seMeta)[,"group",drop=FALSE] %>% data.frame()
plotMat <- metaMat[filter(resTab, P.Value < 0.25)$metabolite,]
pdf("../docs/meta_heatmap_sig.pdf",height = 4, width = 10)
pheatmap(plotMat, scale = "row",
annotation_col = colAnno,
cluster_rows = T, cluster_cols = FALSE,
color = colorRampPalette(c("navy", "white", "firebrick"))(100),
breaks = seq(-3,3,length.out=100),
annotation_colors = annoColor, annotation_names_col = FALSE, fontsize_row = 12)
dev.off()Write CSV output
write_csv2(select(resTab, metabolite, logFC, P.Value, adj.P.Val), "./metabolite_P_values.csv")
write_csv2(as_tibble(metaMat, rownames = "metabolite"), "./metabolite_normalized_abundance.csv")Analysis of the proteomic dataset
seProt <- maeObj[["Proteome"]]
dim(seProt)[1] 2460 12Heatmap visualization
library(pheatmap)
#select top 1000 most variant
colAnno <- colData(seProt)[,c(1:13,19:22)] %>% data.frame()
protMat <- assays(seProt)[["imputed"]]
sds <- genefilter::rowSds(protMat)
protMat <- protMat[order(sds, decreasing = T)[1:1000],]
pheatmap(protMat, show_rownames = FALSE, scale = "row",
annotation_col = colAnno,
clustering_method = "ward.D2")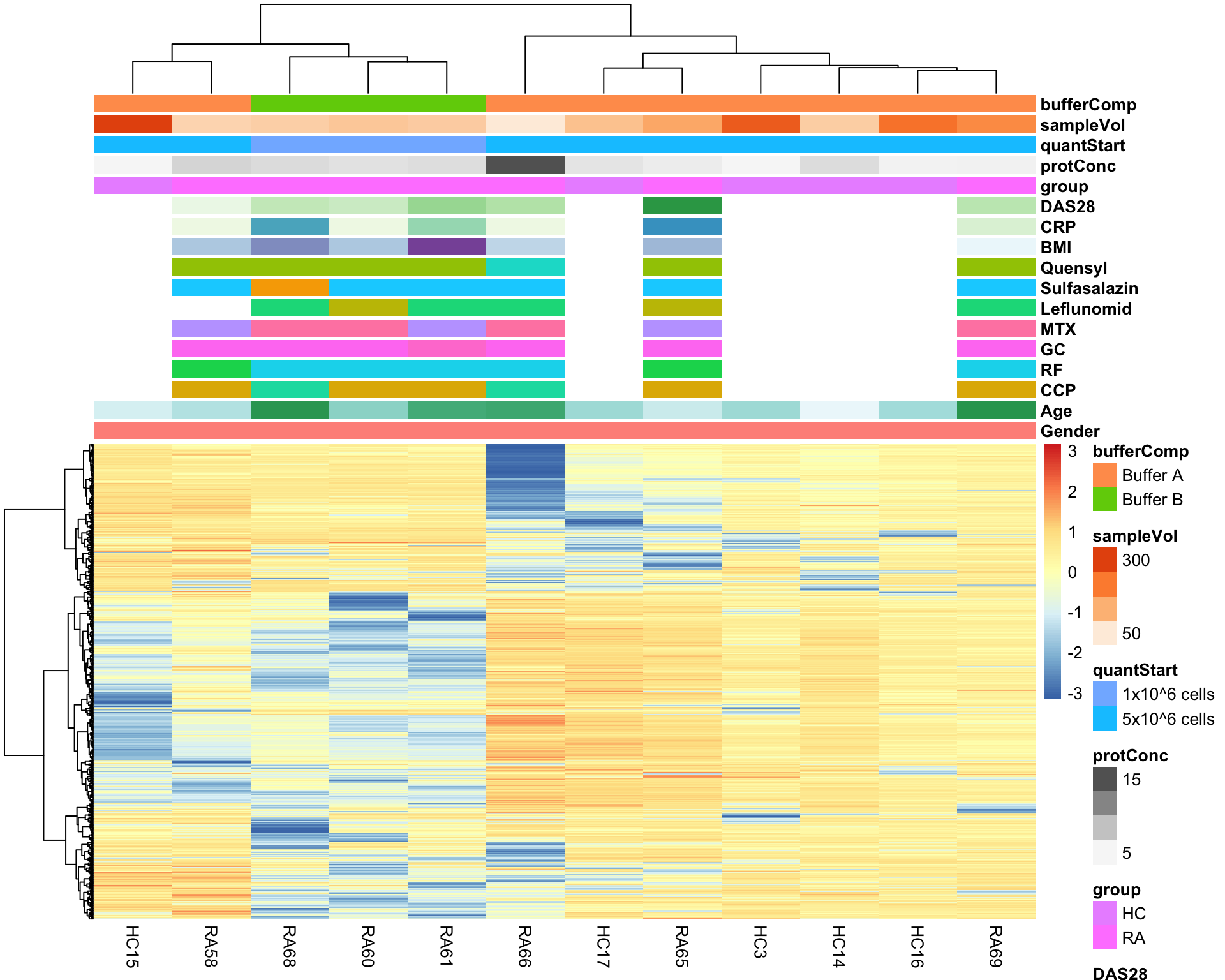
PCA
prRes <- prcomp(t(protMat), scale. = TRUE, center = TRUE)
plotTab <- prRes$x %>% as_tibble(rownames = "sampleID") %>%
left_join(as_tibble(colAnno, rownames = "sampleID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = group, shape = bufferComp)) +
geom_point(size=5) +
ggrepel::geom_text_repel(aes(label = sampleID))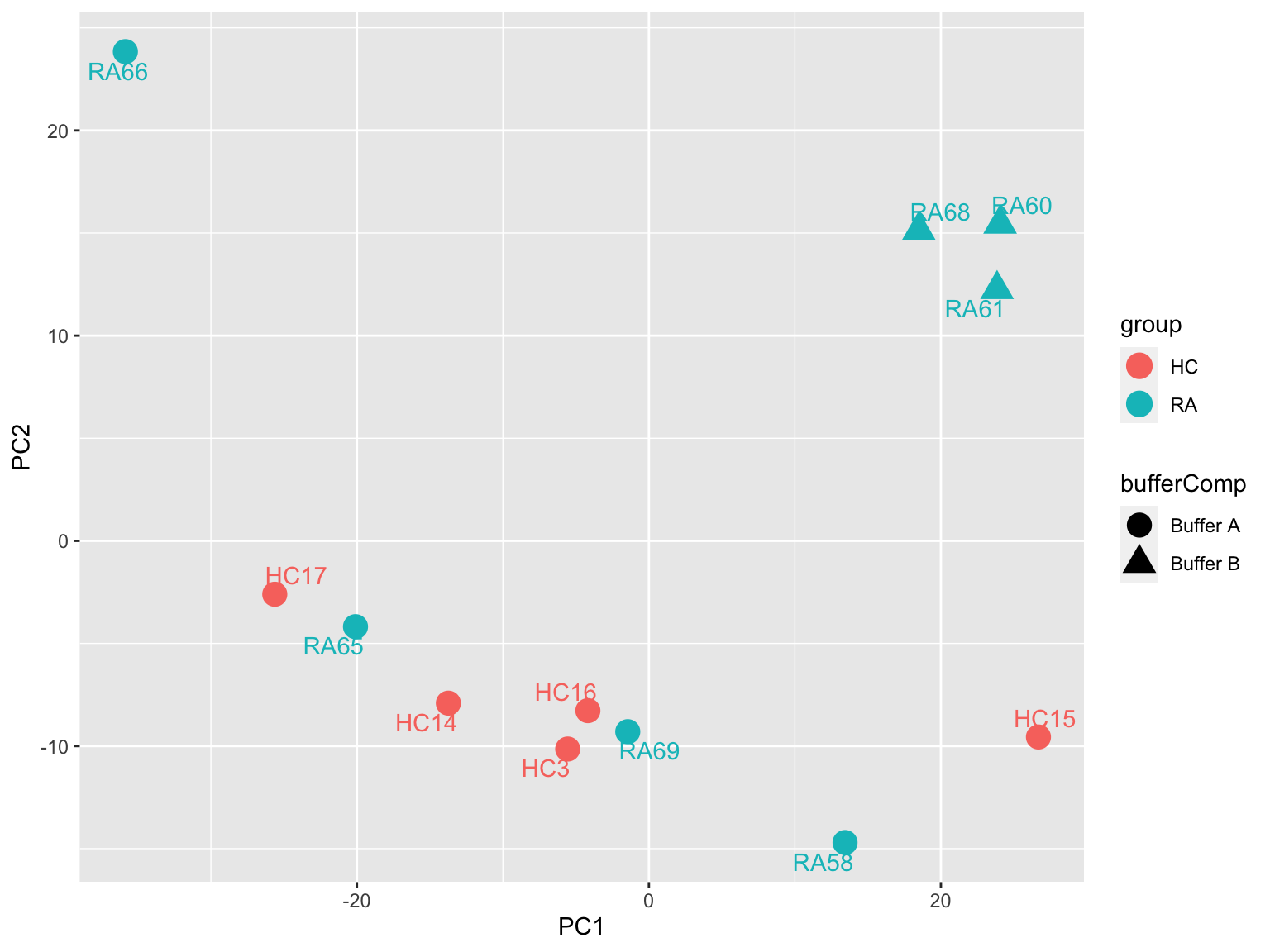
Differential expression using proDA
Differential protein expression using proDA, blocked for buffer condition
library(proDA)
protMat <- assays(seProt)[["norm"]]
designMat <- model.matrix(~ bufferComp + group , colData(seProt))
fit <- proDA(protMat, design = designMat)
resTab <- test_diff(fit, contrast = "groupRA") %>%
arrange(pval) %>%
mutate(symbol = rowData(seProt[name,])$symbol)
resTab_prot <- resTab
Warning: The above code chunk cached its results, but
it won’t be re-run if previous chunks it depends on are updated. If you
need to use caching, it is highly recommended to also set
knitr::opts_chunk$set(autodep = TRUE) at the top of the
file (in a chunk that is not cached). Alternatively, you can customize
the option dependson for each individual chunk that is
cached. Using either autodep or dependson will
remove this warning. See the
knitr cache options for more details.
hist(resTab$pval)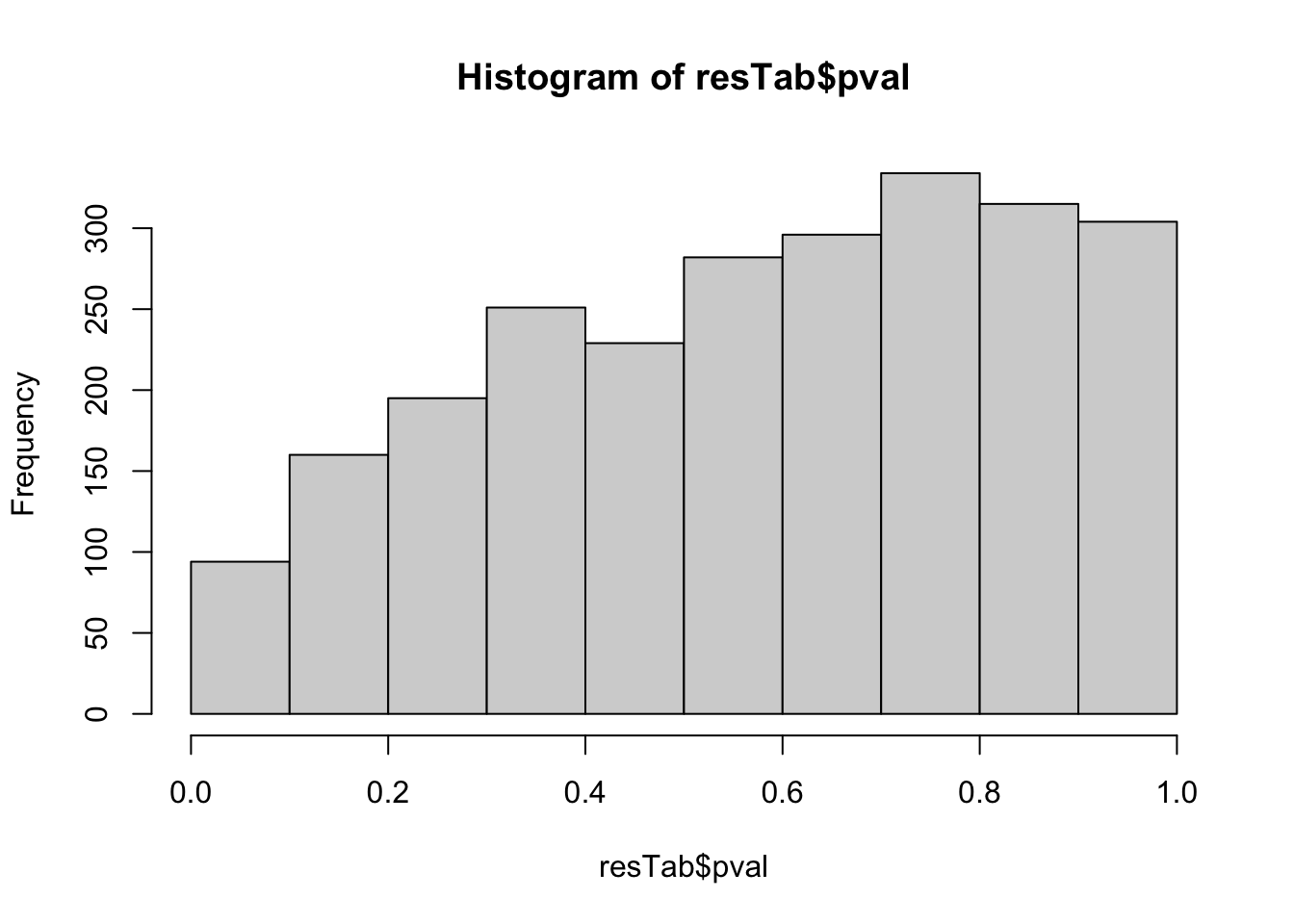 Not strong difference
Not strong difference
Proteins with p-value < 0.05
resTab.sig <- filter(resTab, pval < 0.05)
resTab.sig %>% select(symbol, pval, adj_pval, diff) %>%
mutate_if(is.numeric, formatC, digits=1) %>%
DT::datatable()Plot top 9 examples
pList <- lapply(seq(9), function(i) {
rec <- resTab.sig[i,]
plotTab <- tibble(expr = protMat[rec$name,],
group = seProt$group,
bufferComp = seProt$bufferComp)
ggplot(plotTab, aes(x=group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = group, shape = bufferComp), size=3) +
ggtitle(rec$symbol) +
theme_bw()
})
cowplot::plot_grid(plotlist = pList,ncol=3)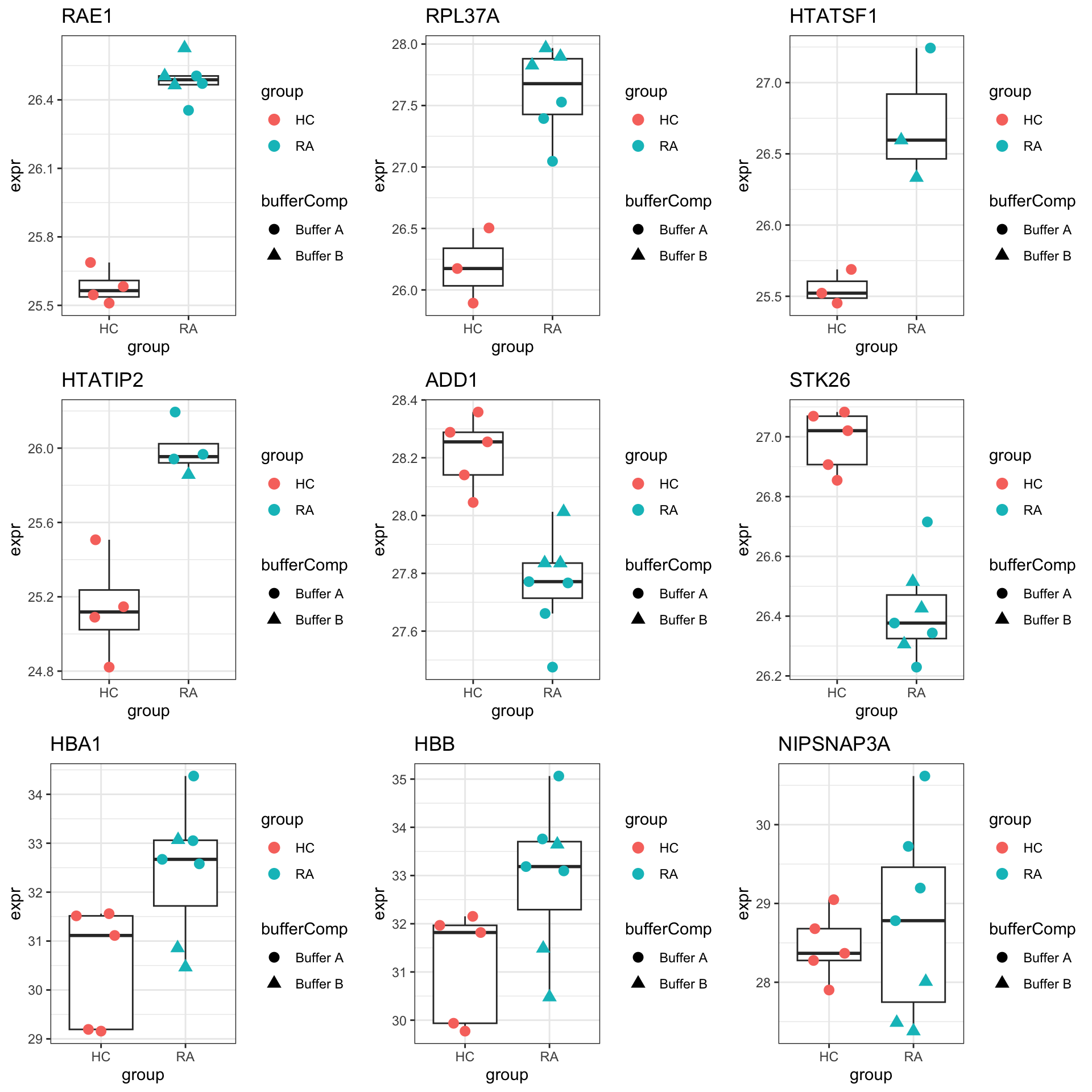
Enrichment analysis
gmts = list(H= "~/EMBL_projects/data/commonFiles/h.all.v6.2.symbols.gmt",
KEGG = "~/EMBL_projects/data/commonFiles/c2.cp.kegg.v6.2.symbols.gmt",
C6 = "~/EMBL_projects/data/commonFiles/c6.all.v6.2.symbols.gmt",
GOBP = "~/EMBL_projects/data/commonFiles/c5.bp.v6.2.symbols.gmt")
inputTab <- resTab %>% filter(pval < 0.1) %>%
distinct(symbol, .keep_all = TRUE) %>%
select(symbol, t_statistic) %>% data.frame() %>% column_to_rownames("symbol")
enRes <- list()
enRes[["Hallmark"]] <- runGSEA(inputTab, gmts$H, "page")
enRes[["KEGG"]] <- runGSEA(inputTab, gmts$KEGG,"page")
enRes[["Perturbation"]] <- runGSEA(inputTab, gmts$C6,"page")
enRes[["GOBP"]] <- runGSEA(inputTab, gmts$GOBP,"page")
p <- plotEnrichmentBar(enRes, pCut =0.05, ifFDR= FALSE)
cowplot::plot_grid(p)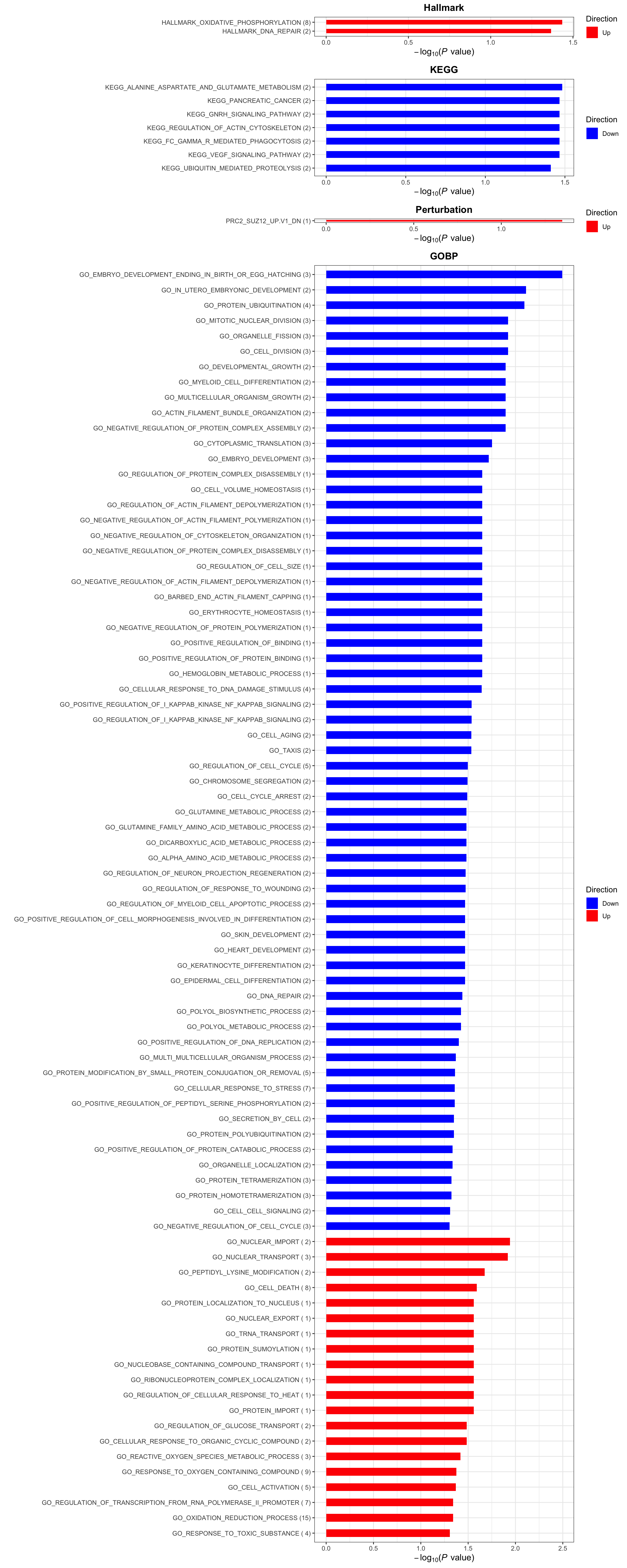
Association between protein expression and enhancer methylation levels
Are there any overlap between differential expressed proteins and differential methylated enhancer CpGs for the same protein?
#get significant protiens
resTab_prot <- resTab_prot %>%
filter(pval < 0.05) %>%
dplyr::rename(fc.prot = diff, p.prot = pval, padj.prot = adj_pval, protID = name) %>%
select(protID, symbol, fc.prot, p.prot, padj.prot)
#get significant enhancer CpGs list
load("../output/resTab_enhancerCpG.RData")
resTab_enhancerCpG <- filter(resTab_enhancerCpG, P.Value < 0.01) %>%
select(-B, -symbol) %>% dplyr::rename(p.cpg = P.Value, padj.cpg = adj.P.Val, fc.cpg = logFC, fc.cpg.beta = logFC_beta, symbol = gene)
#Get DMR regions
dmrRes <- readxl::read_xlsx("../docs/DMR_GeneHancer.xlsx") %>% filter(p.value < 0.01) %>%
dplyr::rename(fc.dmr = estimate, p.dmr = p.value, padj.dmr = p.adjust, symbol = gene) %>%
select(-c(enhancerId, feature, chr, start, end)) %>%
arrange(p.dmr) %>% distinct(symbol,.keep_all = TRUE)
resTab.com <- resTab_enhancerCpG %>% left_join(resTab_prot, by ="symbol") %>%
left_join(dmrRes, by = "symbol") %>%
filter(!is.na(p.prot), !is.na(p.cpg))Overlapped proteins
unique(sort(resTab.com$symbol))[1] "FABP5" "MAP2K1"Only two proteins
Write CSV output
write_csv2(select(resTab, symbol, diff, pval, adj_pval) %>% dplyr::rename(logFC = diff), "../docs/protein_P_values.csv")
write_csv2(as_tibble(protMat, rownames = "id") %>%
mutate(symbol = rowData(seProt)[id,]$symbol) %>%
select(-id), "../docs/protein_normalized_abundance.csv")Analysis of phospho dataset
sePhos <- maeObj[["Phosphoproteome"]]Heatmap visualization
library(pheatmap)
#select top 1000 most variant
colAnno <- colData(seProt)[,c(1:13,19:22)] %>% data.frame()
protMat <- assays(sePhos)[["imputed"]]
sds <- genefilter::rowSds(protMat)
protMat <- protMat[order(sds, decreasing = T)[1:1000],]
pheatmap(protMat, show_rownames = FALSE, scale = "row",
annotation_col = colAnno,
clustering_method = "ward.D2")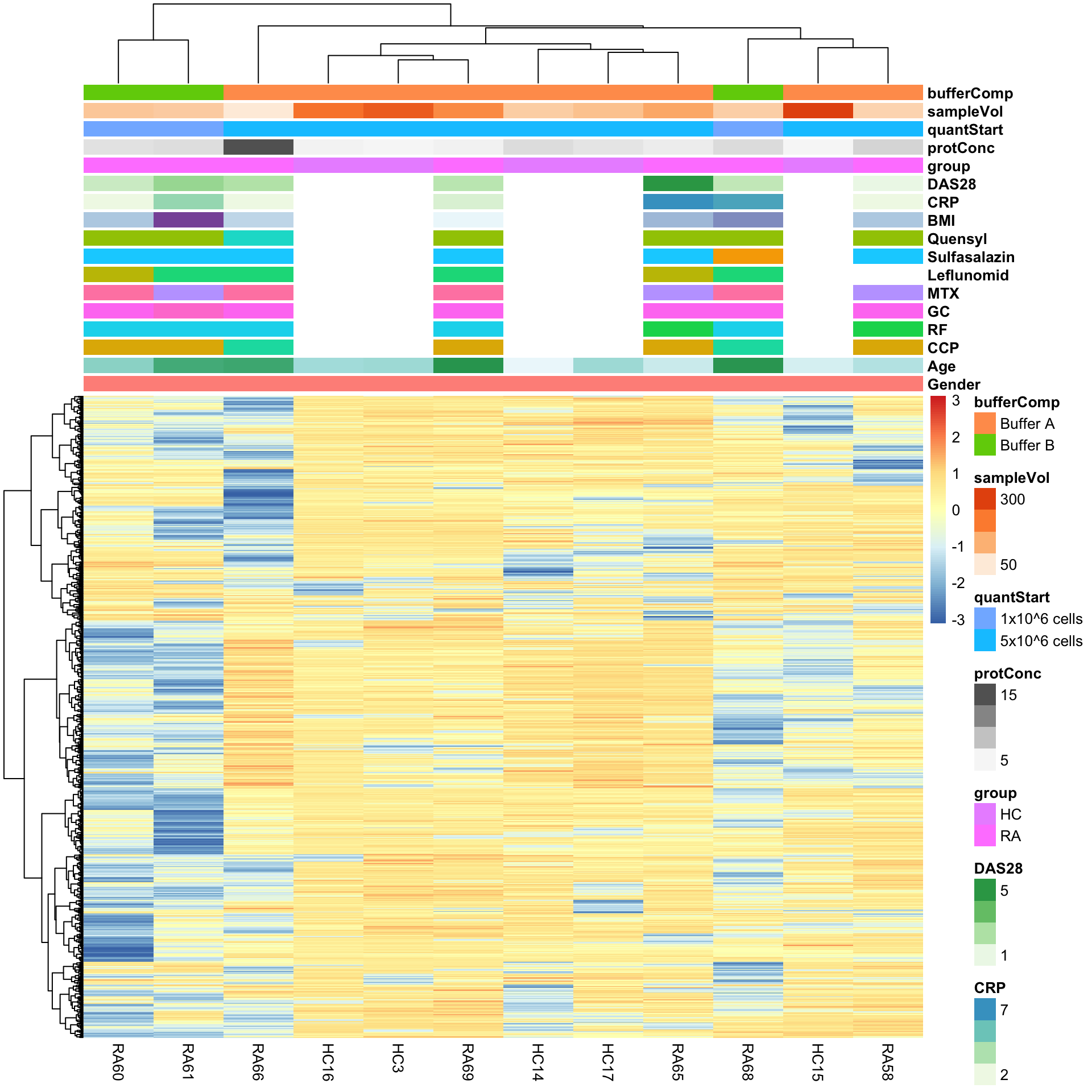
PCA
prRes <- prcomp(t(protMat), scale. = TRUE, center = TRUE)
plotTab <- prRes$x %>% as_tibble(rownames = "sampleID") %>%
left_join(as_tibble(colAnno, rownames = "sampleID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = group, shape = bufferComp)) +
geom_point(size=5) +
ggrepel::geom_text_repel(aes(label = sampleID))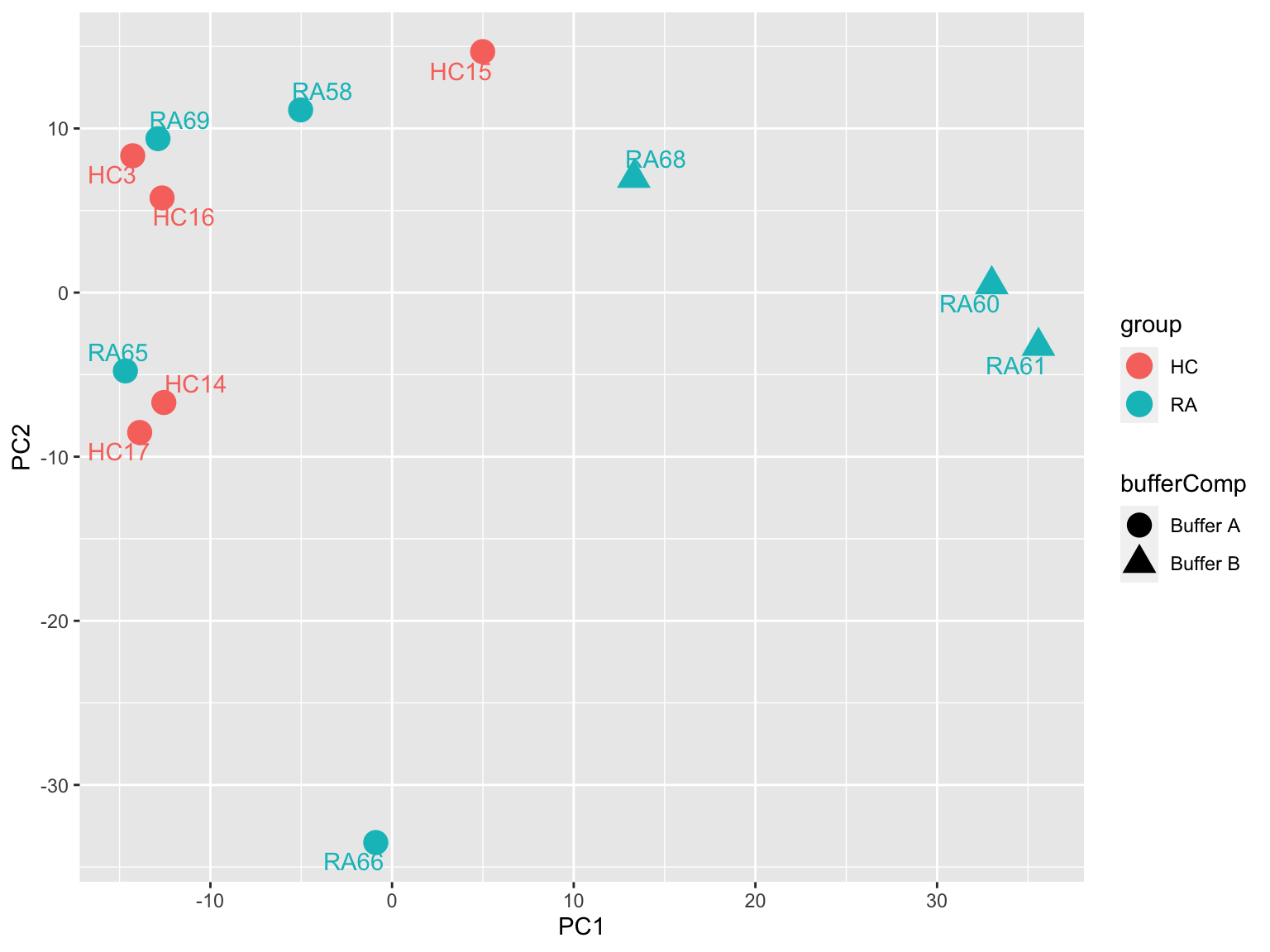 Buffer composition may act as a confounding factor. One sample, RA62 may
be outlier.
Buffer composition may act as a confounding factor. One sample, RA62 may
be outlier.
Differential expression using proDA
Differential protein expression using proDA
protMat <- assays(sePhos)[["norm"]]
designMat <- model.matrix(~ group + bufferComp , colData(sePhos))
fit <- proDA(protMat, design = designMat)
resTab <- test_diff(fit, contrast = "groupRA") %>%
arrange(pval) %>%
mutate(symbol = rowData(sePhos)[name,]$symbol,
site = rowData(sePhos)[name,]$site)
Warning: The above code chunk cached its results, but
it won’t be re-run if previous chunks it depends on are updated. If you
need to use caching, it is highly recommended to also set
knitr::opts_chunk$set(autodep = TRUE) at the top of the
file (in a chunk that is not cached). Alternatively, you can customize
the option dependson for each individual chunk that is
cached. Using either autodep or dependson will
remove this warning. See the
knitr cache options for more details.
hist(resTab$pval)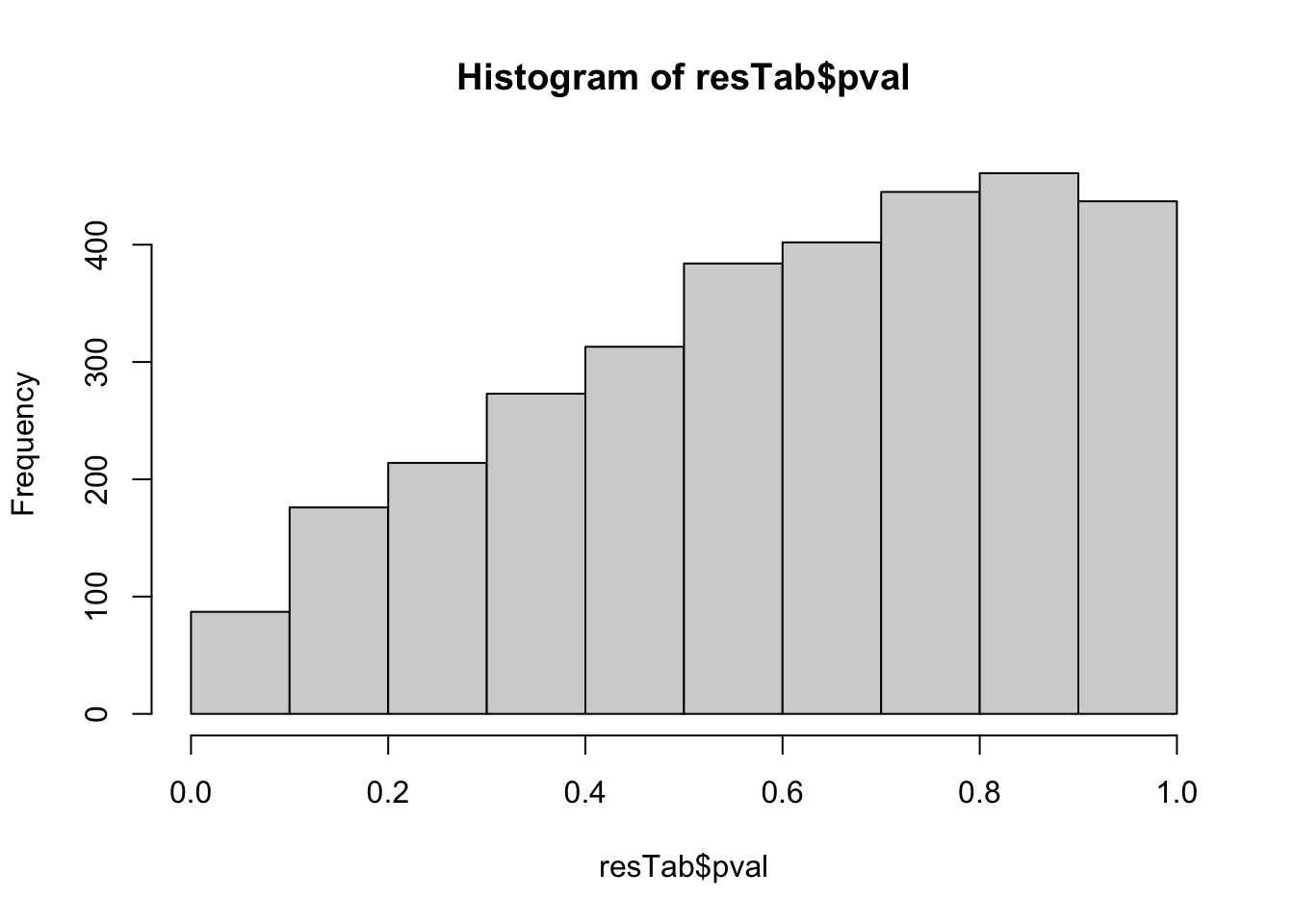 Not strong difference
Not strong difference
Proteins with p-value < 0.05
resTab.sig <- filter(resTab, pval < 0.05)
resTab.sig %>% select(symbol, site, pval, adj_pval, diff) %>%
mutate_if(is.numeric, formatC, digits=1) %>%
DT::datatable()Plot top 9 examples
pList <- lapply(seq(9), function(i) {
rec <- resTab.sig[i,]
plotTab <- tibble(expr = protMat[rec$name,],
group = sePhos$group,
bufferComp = sePhos$bufferComp)
ggplot(plotTab, aes(x=group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = group, shape = bufferComp), size=3) +
ggtitle(rec$site) +
theme_bw()
})
cowplot::plot_grid(plotlist = pList,ncol=3)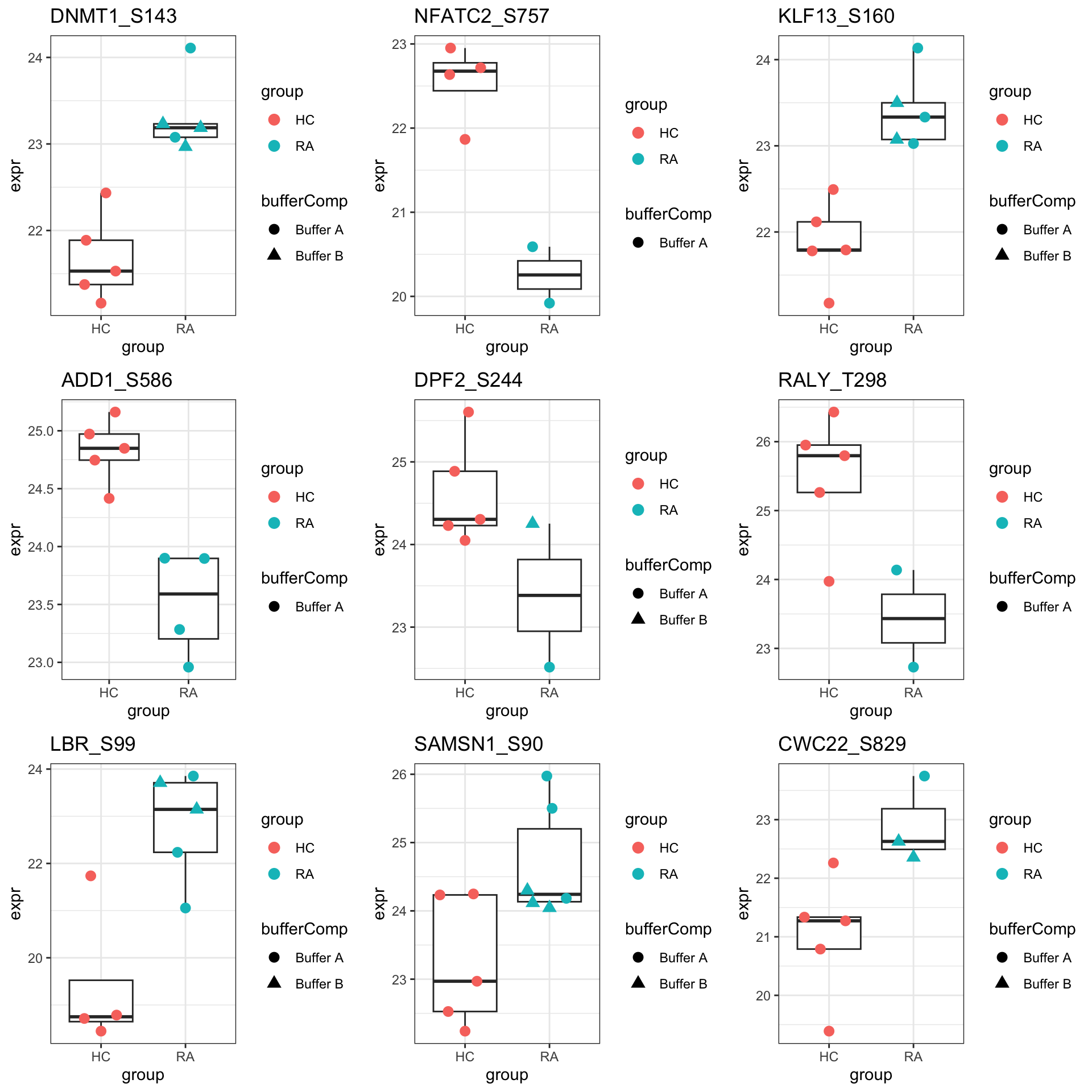
Enrichment analysis
inputTab <- resTab %>% filter(pval < 0.1) %>%
distinct(symbol, .keep_all = TRUE) %>%
select(symbol, t_statistic) %>% data.frame() %>% column_to_rownames("symbol")
enRes <- list()
enRes[["Hallmark"]] <- runGSEA(inputTab, gmts$H, "page")
enRes[["KEGG"]] <- runGSEA(inputTab, gmts$KEGG,"page")
enRes[["Perturbation"]] <- runGSEA(inputTab, gmts$C6,"page")
enRes[["GOBP"]] <- runGSEA(inputTab, gmts$GOBP,"page")
p <- plotEnrichmentBar(enRes, pCut =0.05, ifFDR= FALSE)[1] "No sets passed the criteria"cowplot::plot_grid(p)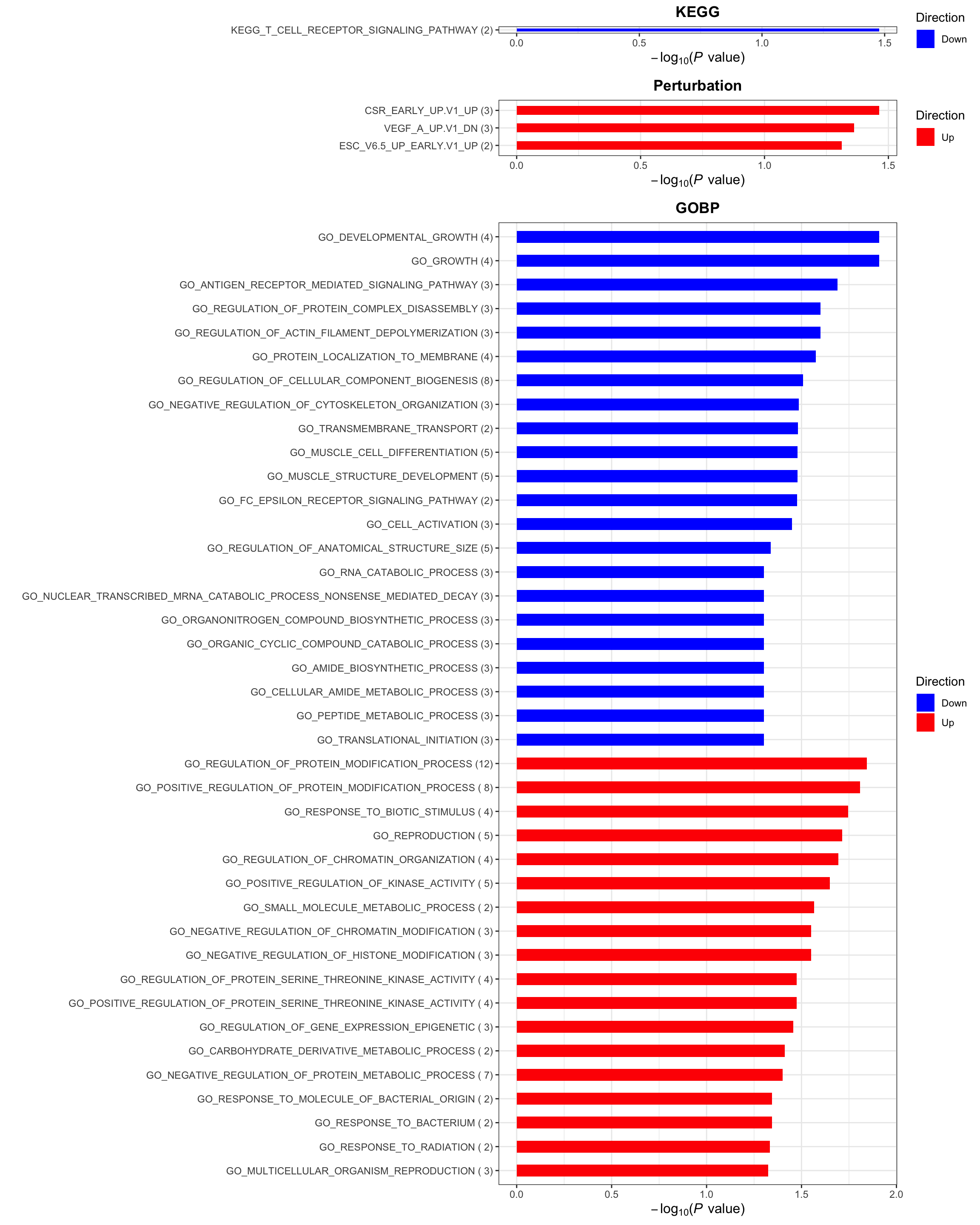
Write CSV output
write_csv2(select(resTab, symbol, site, diff, pval, adj_pval) %>% dplyr::rename(logFC = diff), "../docs/phos_P_values.csv")
write_csv2(as_tibble(protMat, rownames = "id") %>%
mutate(symbol = rowData(sePhos)[id,]$symbol,
site = rowData(sePhos)[id,]$site) %>%
select(-id), "../docs/phosphorylation_normalized_abundance.csv")Kinase activity analysis using decoupler
library(PHONEMeS)
library(decoupleR)#get decoupler network
decoupler_network <- phonemesPKN %>%
dplyr::rename("mor" = interaction) %>%
tibble::add_column("likelihood" = 1)
#define decoupler input
decoupler_input <- resTab %>%
dplyr::filter(site %in% decoupler_network$target) %>%
distinct(site, .keep_all = TRUE) %>%
tibble::column_to_rownames("site") %>%
dplyr::select(t_statistic)
#filter deoupler network
decoupler_network <- decoupleR::intersect_regulons(mat = decoupler_input,
network = decoupler_network,
.source = source,
.target = target,
minsize = 5)
#remove overlapped regulons
correlated_regulons <- decoupleR::check_corr(decoupler_network) %>%
dplyr::filter(correlation >= 0.9)
decoupler_network <- decoupler_network %>%
dplyr::filter(!source %in% correlated_regulons$source.2)
# run mlm to estimate kinase activities
kinase_activity <- decoupleR::run_mlm(mat = decoupler_input,
network = decoupler_network)
head(kinase_activity)# A tibble: 6 × 5
statistic source condition score p_value
<chr> <chr> <chr> <dbl> <dbl>
1 mlm SGK1 t_statistic 1.74 0.0832
2 mlm CHEK1 t_statistic -0.244 0.807
3 mlm AURKA t_statistic -1.04 0.301
4 mlm PDPK1 t_statistic -1.04 0.300
5 mlm PRKY t_statistic -0.0912 0.927
6 mlm ROCK2 t_statistic -0.593 0.554 Analysis of phospho dataset (normalized by protein expression)
sePhos <- maeObj[["PhosRatio"]]
sePhos$group <- maeObj[,sePhos$colID]$group
#colData(sePhos) <- colData(maeObj[,colnames(sePhos)])Differential expression using proDA
Differential protein expression using proDA
protMat <- assays(sePhos)[["ratio"]]
designMat <- model.matrix(~ group + bufferComp, colData(sePhos))
fit <- proDA(protMat, design = designMat)
resTab <- test_diff(fit, contrast = "groupRA") %>%
arrange(pval) %>%
mutate(symbol = rowData(sePhos)[name,]$symbol,
site = rowData(sePhos)[name,]$site)
Warning: The above code chunk cached its results, but
it won’t be re-run if previous chunks it depends on are updated. If you
need to use caching, it is highly recommended to also set
knitr::opts_chunk$set(autodep = TRUE) at the top of the
file (in a chunk that is not cached). Alternatively, you can customize
the option dependson for each individual chunk that is
cached. Using either autodep or dependson will
remove this warning. See the
knitr cache options for more details.
hist(resTab$pval)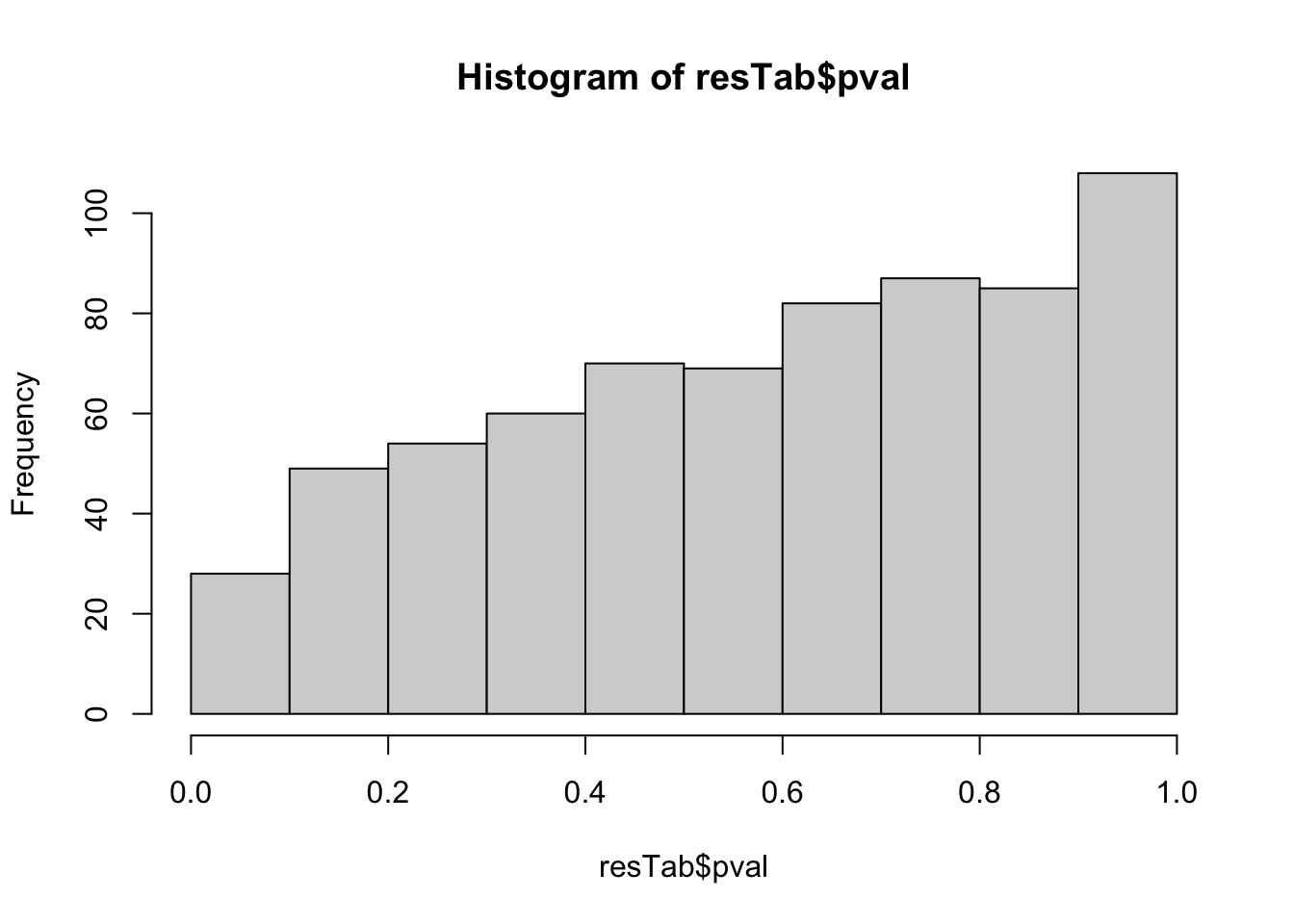 Not strong difference
Not strong difference
Proteins with p-value < 0.05
resTab.sig <- filter(resTab, pval < 0.05)
resTab.sig %>% select(symbol, site, pval, adj_pval, diff) %>%
mutate_if(is.numeric, formatC, digits=1) %>%
DT::datatable()Plot top 9 examples
pList <- lapply(seq(9), function(i) {
rec <- resTab.sig[i,]
plotTab <- tibble(expr = protMat[rec$name,],
group = sePhos$group,
bufferComp = sePhos$bufferComp)
ggplot(plotTab, aes(x=group, y=expr)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = group, shape = bufferComp), size=3) +
ggtitle(rec$site) +
theme_bw()
})
cowplot::plot_grid(plotlist = pList,ncol=3)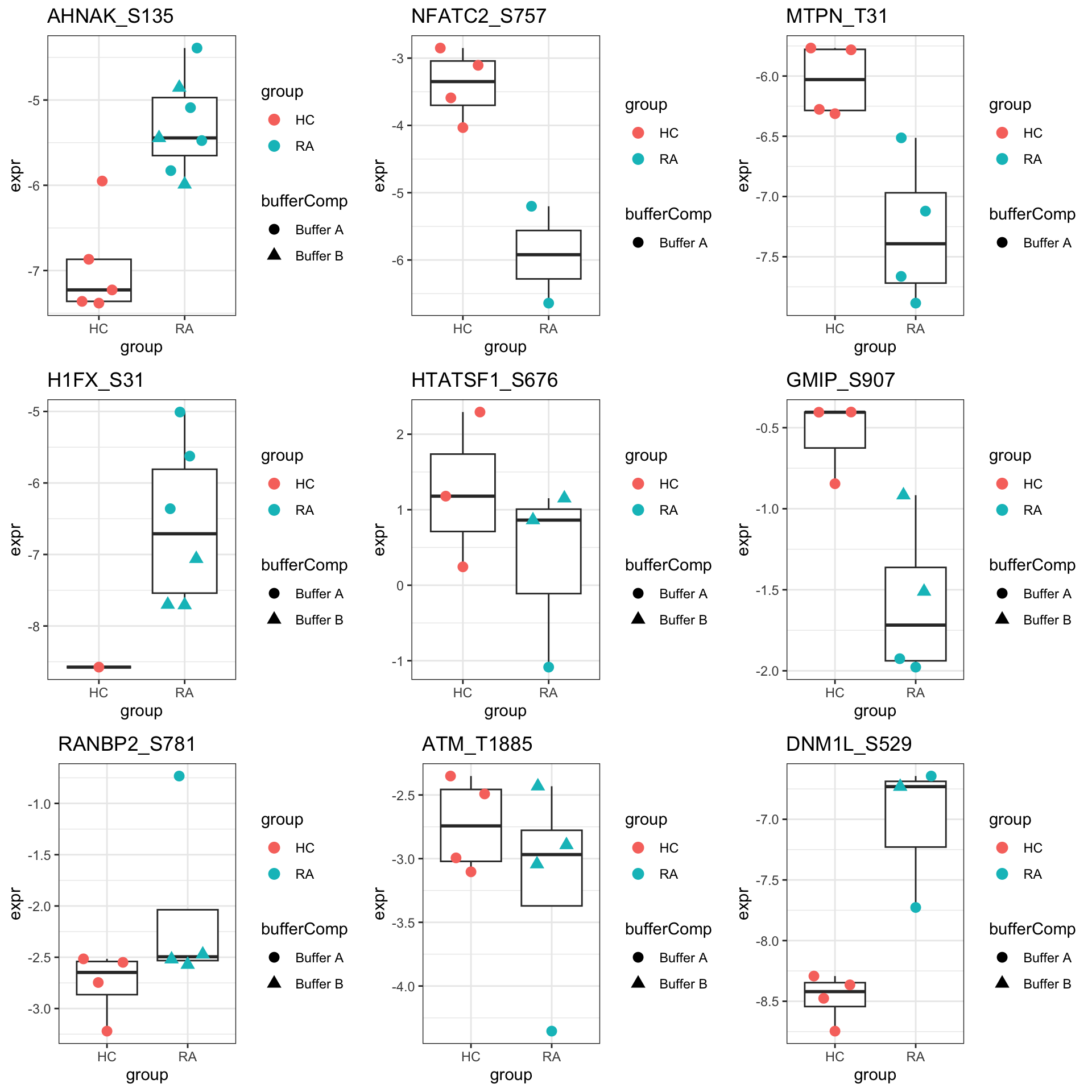
Enrichment analysis
inputTab <- resTab %>% filter(pval < 0.1) %>%
distinct(symbol, .keep_all = TRUE) %>%
select(symbol, t_statistic) %>% data.frame() %>% column_to_rownames("symbol")
enRes <- list()
enRes[["Hallmark"]] <- runGSEA(inputTab, gmts$H, "page")
enRes[["KEGG"]] <- runGSEA(inputTab, gmts$KEGG,"page")
enRes[["Perturbation"]] <- runGSEA(inputTab, gmts$C6,"page")
enRes[["GOBP"]] <- runGSEA(inputTab, gmts$GOBP,"page")
p <- plotEnrichmentBar(enRes, pCut =0.05, ifFDR= FALSE)[1] "No sets passed the criteria"cowplot::plot_grid(p)
Write CSV output
write_csv2(select(resTab, symbol, site, diff, pval, adj_pval) %>% dplyr::rename(logFC = diff), "../docs/phos_P_values_protNorm.csv")
write_csv2(as_tibble(protMat, rownames = "id") %>%
mutate(symbol = rowData(sePhos)[id,]$symbol,
site = rowData(sePhos)[id,]$site) %>%
select(-id), "../docs/phosphorylation_ProtNormalized_abundance.csv")Kinase activity analysis using decoupler
#get decoupler network
decoupler_network <- phonemesPKN %>%
dplyr::rename("mor" = interaction) %>%
tibble::add_column("likelihood" = 1)
#define decoupler input
decoupler_input <- resTab %>%
dplyr::filter(site %in% decoupler_network$target) %>%
distinct(site, .keep_all = TRUE) %>%
tibble::column_to_rownames("site") %>%
dplyr::select(t_statistic)
#filter deoupler network
decoupler_network <- decoupleR::intersect_regulons(mat = decoupler_input,
network = decoupler_network,
.source = source,
.target = target,
minsize = 5)
#remove overlapped regulons
correlated_regulons <- decoupleR::check_corr(decoupler_network) %>%
dplyr::filter(correlation >= 0.9)
decoupler_network <- decoupler_network %>%
dplyr::filter(!source %in% correlated_regulons$source.2)
# run mlm to estimate kinase activities
kinase_activity <- decoupleR::run_mlm(mat = decoupler_input,
network = decoupler_network)
head(kinase_activity)# A tibble: 6 × 5
statistic source condition score p_value
<chr> <chr> <chr> <dbl> <dbl>
1 mlm PRKY t_statistic 0.145 0.885
2 mlm PRKCG t_statistic -0.464 0.643
3 mlm PRKCB t_statistic 0.198 0.843
4 mlm CDK1 t_statistic 0.384 0.701
5 mlm PRKCA t_statistic 0.108 0.914
6 mlm PRKACA t_statistic -0.120 0.905FACS data
seFacs <- maeObj[["FACS"]]
facsTab <- sumToTidy(seFacs, rowID = "id", colID = "sampleID")
facsMat <- assay(seFacs)Vsn
facsMat.vst <- vsn::justvsn(facsMat)
#meanSdPlot(facsMat.vst)PCA
Perform PCA
pcMat <- facsMat.vst
pcMat <- pcMat[complete.cases(pcMat),]
pcRes <- prcomp(t(pcMat), scale. = TRUE, center = TRUE)$x
plotTab <- as_tibble(pcRes, rownames = "colID") %>%
left_join(as.tibble(colData(seFacs), rownames = "colID"))
ggplot(plotTab, aes(x=PC1, y=PC2, col = group, label = sampleID)) +
geom_point() +
ggrepel::geom_text_repel()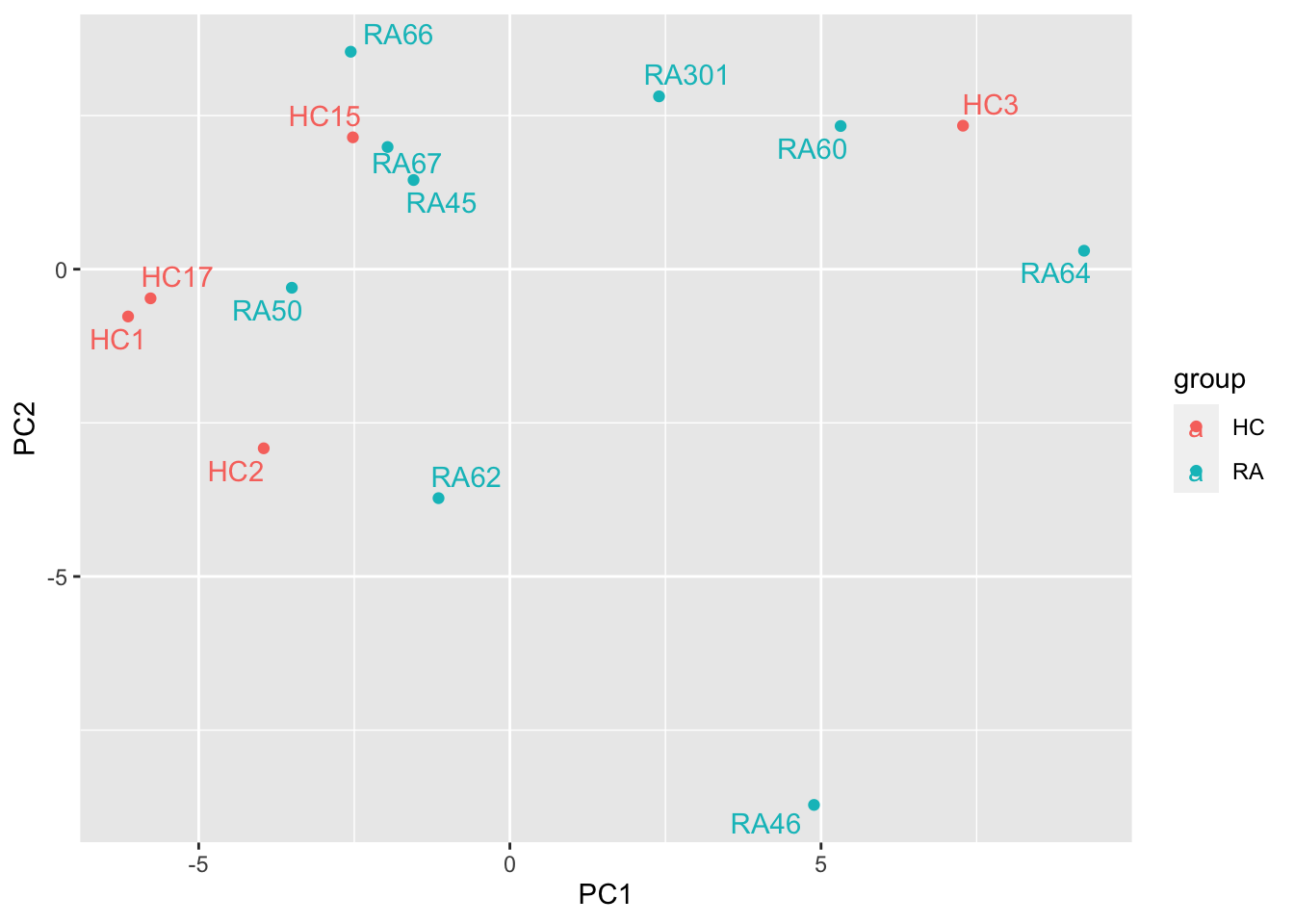
Differential test
designMat <- model.matrix(~group, data = colData(seFacs))
lmFit <- lmFit(facsMat.vst, design = designMat)
fit2 <- eBayes(lmFit)
resTab <- topTable(fit2, number = Inf) %>%
as_tibble(rownames = "id") %>%
mutate(feature = rowData(seFacs[id,])$feature)
hist(resTab$P.Value)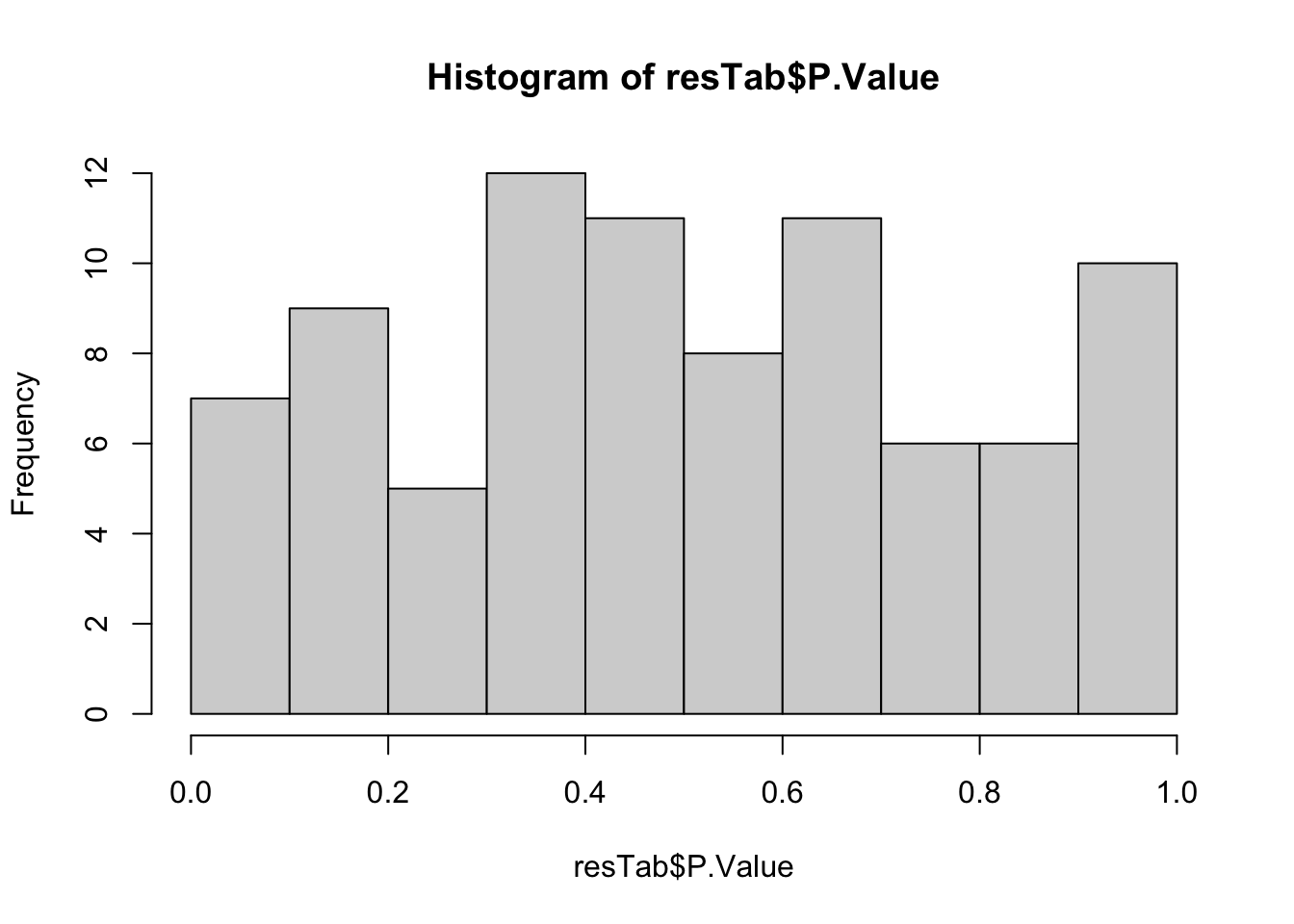
head(resTab)# A tibble: 6 × 8
id logFC AveExpr t P.Value adj.P.Val B feature
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 f36 0.910 9.49 2.30 0.0351 0.910 -4.51 effector memory PD1
2 f29 -0.910 5.07 -2.18 0.0444 0.910 -4.52 effector memory CTLA4
3 f49 -0.634 5.42 -2.06 0.0556 0.910 -4.53 effector TEMRA CTLA4
4 f56 0.945 9.08 2.02 0.0604 0.910 -4.53 effector TEMRA PD1
5 f74 -0.826 13.1 -1.91 0.0746 0.910 -4.54 naive LDHA
6 f17 -0.932 13.3 -1.86 0.0807 0.910 -4.54 central memory LDHA Plot significant associations
pList <- lapply(seq(nrow(filter(resTab, P.Value <= 0.05))), function(i) {
rec <- resTab[i,]
plotTab <- filter(facsTab, id == rec$id)
ggplot(plotTab, aes(x=group, y=count, label = sampleID)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(color = group), size=3) +
ggtitle(sprintf("%s (P=%s)",rec$feature,formatC(rec$P.Value,digits = 2))) +
ggrepel::geom_text_repel() +
theme_bw() +
theme(legend.position = "none")
})
cowplot::plot_grid(plotlist = pList,ncol=3)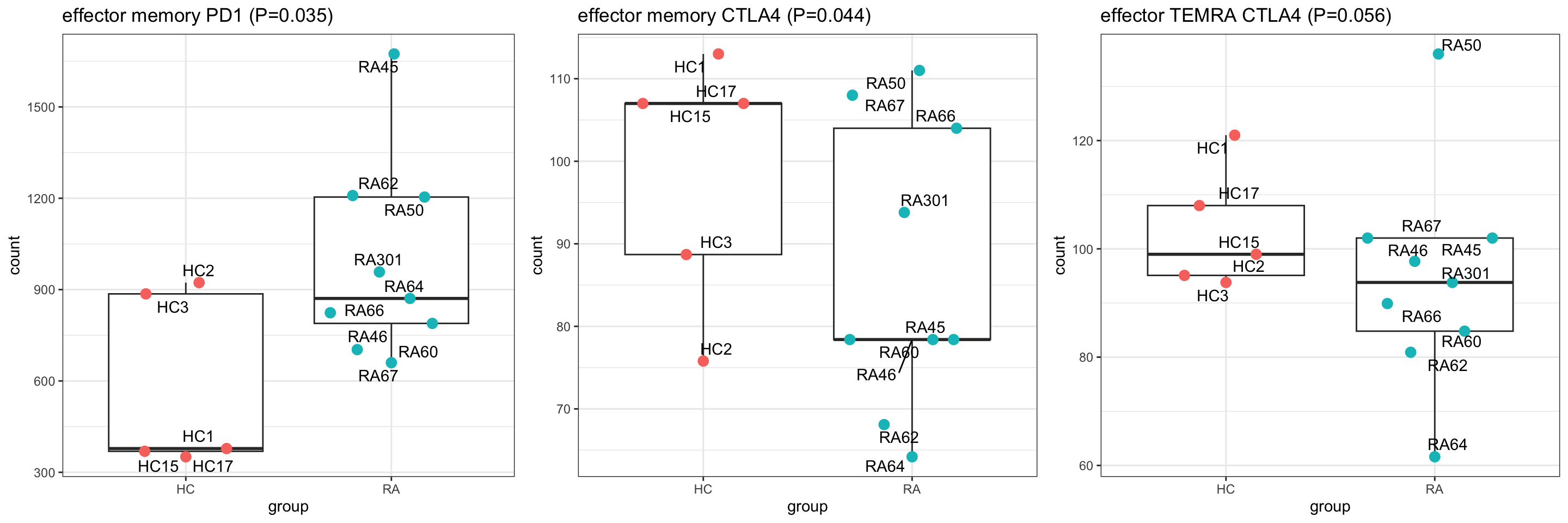
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] decoupleR_2.2.2 PHONEMeS_2.0.1
[3] piano_2.12.0 pheatmap_1.0.12
[5] limma_3.52.2 forcats_0.5.1
[7] stringr_1.4.1 dplyr_1.1.4.9000
[9] purrr_0.3.4 readr_2.1.2
[11] tidyr_1.2.0 tibble_3.2.1
[13] ggplot2_3.4.1 tidyverse_1.3.2
[15] proDA_1.10.0 MultiAssayExperiment_1.22.0
[17] SummarizedExperiment_1.26.1 Biobase_2.56.0
[19] GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[21] IRanges_2.30.0 S4Vectors_0.34.0
[23] BiocGenerics_0.42.0 MatrixGenerics_1.8.1
[25] matrixStats_0.62.0 jyluMisc_0.1.5
loaded via a namespace (and not attached):
[1] utf8_1.2.4 shinydashboard_0.7.2 tidyselect_1.2.1
[4] RSQLite_2.2.15 AnnotationDbi_1.58.0 htmlwidgets_1.5.4
[7] grid_4.2.0 BiocParallel_1.30.4 maxstat_0.7-25
[10] munsell_0.5.0 preprocessCore_1.58.0 ragg_1.2.2
[13] codetools_0.2-18 DT_0.23 withr_3.0.0
[16] colorspace_2.0-3 highr_0.9 knitr_1.39
[19] rstudioapi_0.13 ggsignif_0.6.3 labeling_0.4.2
[22] git2r_0.30.1 slam_0.1-50 GenomeInfoDbData_1.2.8
[25] KMsurv_0.1-5 bit64_4.0.5 farver_2.1.1
[28] rprojroot_2.0.3 vctrs_0.6.5 generics_0.1.3
[31] TH.data_1.1-1 xfun_0.31 sets_1.0-21
[34] R6_2.5.1 ggbeeswarm_0.6.0 locfit_1.5-9.6
[37] bitops_1.0-7 cachem_1.0.6 fgsea_1.22.0
[40] DelayedArray_0.22.0 assertthat_0.2.1 vroom_1.5.7
[43] promises_1.2.0.1 scales_1.2.0 multcomp_1.4-26
[46] googlesheets4_1.0.0 beeswarm_0.4.0 gtable_0.3.0
[49] extraDistr_1.9.1 affy_1.74.0 sva_3.44.0
[52] sandwich_3.0-2 workflowr_1.7.0 rlang_1.1.3
[55] genefilter_1.78.0 systemfonts_1.0.4 splines_4.2.0
[58] rstatix_0.7.0 gargle_1.2.0 broom_1.0.0
[61] BiocManager_1.30.18 yaml_2.3.5 abind_1.4-5
[64] modelr_0.1.8 crosstalk_1.2.0 backports_1.4.1
[67] httpuv_1.6.6 tools_4.2.0 relations_0.6-12
[70] affyio_1.66.0 ellipsis_0.3.2 gplots_3.1.3
[73] RColorBrewer_1.1-3 jquerylib_0.1.4 Rcpp_1.0.11
[76] visNetwork_2.1.0 zlibbioc_1.42.0 RCurl_1.98-1.7
[79] ggpubr_0.4.0 cowplot_1.1.1 zoo_1.8-10
[82] ggrepel_0.9.1 haven_2.5.0 cluster_2.1.3
[85] exactRankTests_0.8-35 fs_1.5.2 magrittr_2.0.3
[88] data.table_1.14.10 reprex_2.0.1 survminer_0.4.9
[91] googledrive_2.0.0 mvtnorm_1.1-3 hms_1.1.1
[94] shinyjs_2.1.0 mime_0.12 evaluate_0.15
[97] xtable_1.8-4 XML_3.99-0.10 readxl_1.4.0
[100] gridExtra_2.3 compiler_4.2.0 KernSmooth_2.23-20
[103] crayon_1.5.2 htmltools_0.5.4 mgcv_1.8-40
[106] later_1.3.0 tzdb_0.3.0 lubridate_1.8.0
[109] DBI_1.1.3 dbplyr_2.2.1 MASS_7.3-58
[112] Matrix_1.5-4 car_3.1-0 cli_3.6.2
[115] vsn_3.64.0 marray_1.74.0 parallel_4.2.0
[118] igraph_1.3.4 pkgconfig_2.0.3 km.ci_0.5-6
[121] xml2_1.3.3 annotate_1.74.0 vipor_0.4.5
[124] bslib_0.4.1 XVector_0.36.0 drc_3.0-1
[127] rvest_1.0.2 digest_0.6.30 Biostrings_2.64.0
[130] rmarkdown_2.14 cellranger_1.1.0 fastmatch_1.1-3
[133] survMisc_0.5.6 edgeR_3.38.1 shiny_1.7.4
[136] gtools_3.9.3 lifecycle_1.0.4 nlme_3.1-158
[139] jsonlite_1.8.3 carData_3.0-5 fansi_1.0.6
[142] pillar_1.9.0 lattice_0.20-45 KEGGREST_1.36.3
[145] fastmap_1.1.0 httr_1.4.3 plotrix_3.8-2
[148] survival_3.4-0 glue_1.7.0 png_0.1-7
[151] bit_4.0.4 stringi_1.7.8 sass_0.4.2
[154] blob_1.2.3 textshaping_0.3.6 caTools_1.18.2
[157] memoise_2.0.1