pkgNeed = c("knitr", "dplyr", "ggplot2", "tidyr", "purrr", "devtools", "reshape2", "stringr",
"terrainr", "imagefx", "dill/beyonce", "EBImage")
pkgInst = installed.packages()[, 1]
if (!("BiocManager" %in% pkgInst)) install.packages("BiocManager")
todo = !(stringr::str_split_i(pkgNeed, "/", -1) %in% pkgInst)
if (any(todo)) BiocManager::install(pkgNeed[todo])Working with Image Data in R
Based on Chapter 11 of “Modern Statistics for Modern Biology”
2023-03-23
Setup and resources
GitHub repo: please clone
https://github.com/wolfganghuber/WorkingWithImageData
Rendered version of the demo:
https://www.huber.embl.de/users/whuber/2304-Imaging-Data-in-R/demo.html
Talk slides (PDF):
https://www.huber.embl.de/users/whuber/2304-Imaging-Data-in-R/talk.pdf
Additional resources:
https://www.huber.embl.de/users/whuber/2304-Imaging-Data-in-R/resources/
ffmpeg:
https://ffmpeg.org or from your package manager
Install the needed R packages
Download example data files. Please setwd to the top directory of the WorkingWithImageData repository
url = "https://www.huber.embl.de/users/whuber/2304-Imaging-Data-in-R/resources/"
files = c("eakKfY5aHmY.mp4", "fvec.RData", "murmuration-flow.mp4", sprintf("tile%03d.tiff", 1:4))
dest = "resources"
if (!dir.exists(dest))
dir.create(dest)
for (f in files) {
d = file.path(dest, f)
if (!file.exists(d))
download.file(paste0(url, f), d)
}Reading and displaying an image
Algebraic computations
x = 1 - idi
display(x)
Convert into a greyscale image
Histogram
hist(idigr)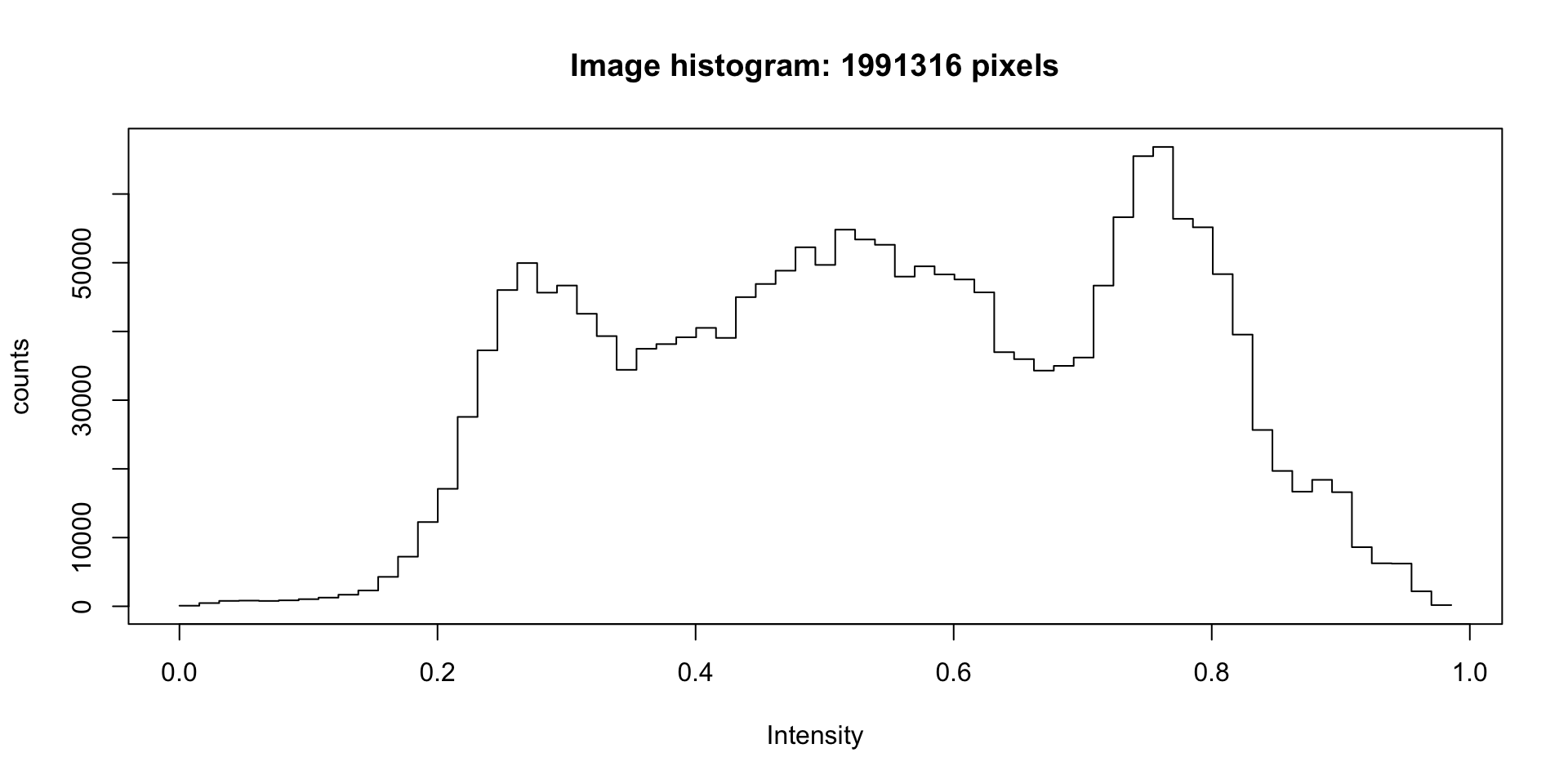
Algebraic computations
x = idigr * 2
display(x)
Computations. An image is just an array
x = idigr ^ (1/3)
display(x)
Thresholding
Transpose
Rotate
Translate
Flip: vertical reflection
Flop: horizontal reflection
Subsetting (‘cropping’)
m = idi[800:1000, 420:560, ]
display(m)
Stitching
files = dir("resources", pattern = "^tile.*.tiff$", full.names = TRUE)
files[1] "resources/tile001.tiff" "resources/tile002.tiff" "resources/tile003.tiff"
[4] "resources/tile004.tiff"tiles = lapply(files, readImage)
tiles[[1]]Image
colorMode : Color
storage.mode : double
dim : 1920 1299 3
frames.total : 3
frames.render: 1
imageData(object)[1:5,1:6,1]
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353
[2,] 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353
[3,] 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353
[4,] 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353
[5,] 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353 0.9882353for (x in tiles) display(x)



Thanks to Joe Davies (@joewdavies) for providing the image.
sx = dim(tiles[[1]])[1] # in practice, look at all tiles and do the gymnastics as necessary
sy = dim(tiles[[1]])[2]
combined = Image(NA_real_, dim = c(2 * sx, 2 * sy, 3), colormode = "color")
combined[ 1:sx , 1:sy, ] = tiles[[1]]
combined[(sx+1):(2*sx), 1:sy, ] = tiles[[2]]
combined[ 1:sx , (sy+1):(2*sy), ] = tiles[[3]]
combined[(sx+1):(2*sx), (sy+1):(2*sy), ] = tiles[[4]]
display(combined)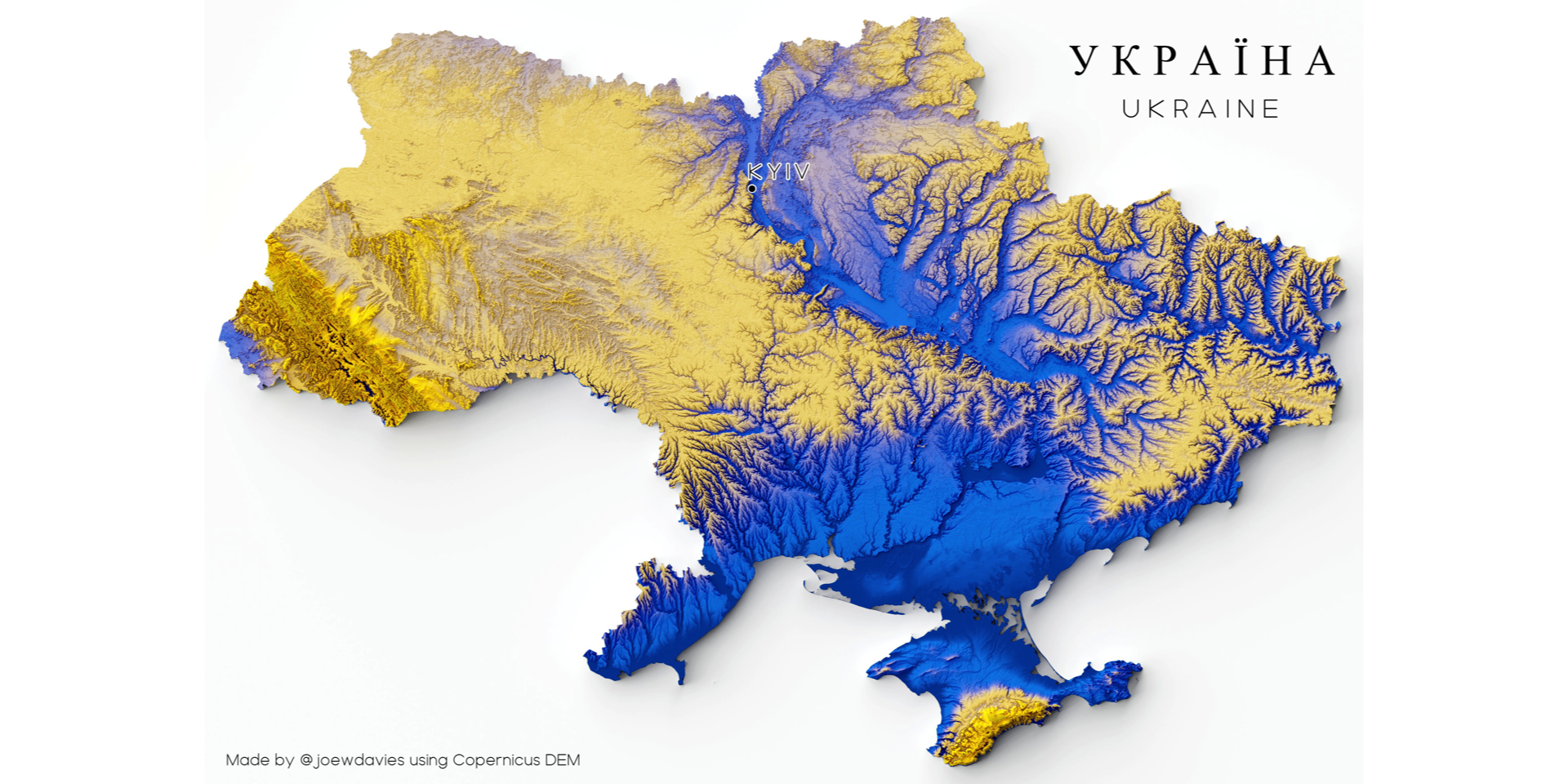
Integration with base R, dplyr, ggplot2
borscz = readImage("fig/borscz_1.jpg")image(borscz)
Credit: https://en.wikipedia.org/wiki/Borscht CC BY 2.0 Liz West from Boxborough, MA
library("ggplot2")
library("dplyr")
library("reshape2") # for melt
library("terrainr") # for geom_spatial_rgb
# Convert 3d array (x * y * RGB-colors) into a tidy dataframe, one row per pixel
array2df = function(x)
melt(x[,,1]) |>
full_join(melt(x[,,2]), by = c("Var1", "Var2")) |>
full_join(melt(x[,,3]), by = c("Var1", "Var2")) |>
`colnames<-`(c("x", "y", "r", "g", "b"))
borsczdf = array2df(borscz) ggplot(borsczdf, aes(x = x, y = -y, r = r, g = g, b = b)) +
geom_spatial_rgb()
Segmentation, object feature extraction and classification
Read an image
grus = readImage("fig/Grus_grus_flocks.jpg") # common crane (Kranich, кран). Source: Wikipedia
display(grus)
Intensity histogram, by color (RGB)
hist(grus)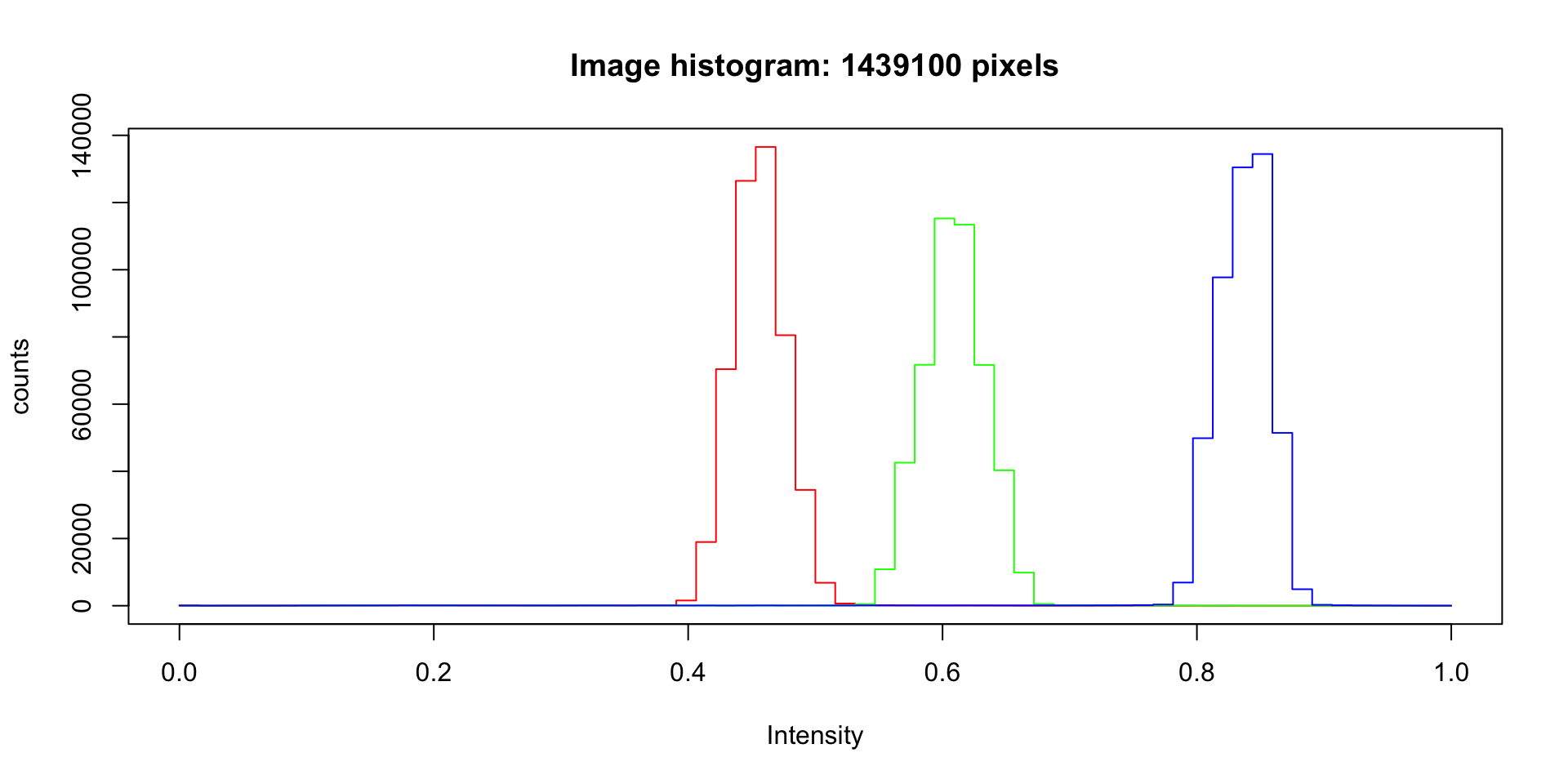
Very simple segmentation: every pixel that’s not very blue is a bird pixel
Find connected components. Intention: each is a separate object of interest (i.e., a bird)
connCompID = bwlabel(grus_fg)
display(colorLabels(connCompID))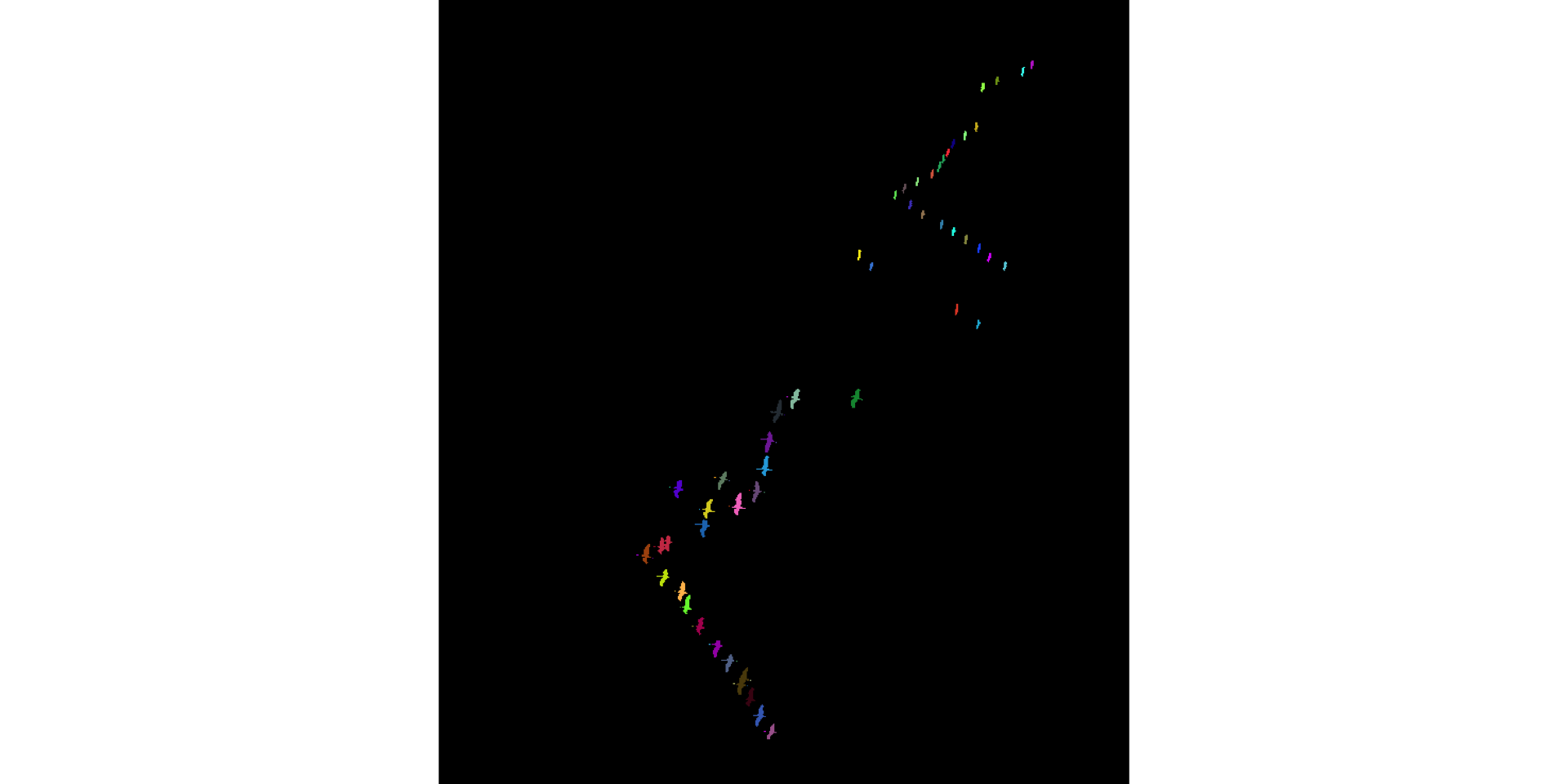
Some heads are cut off due to misclassification of bright-colored neck as sky \(\to\) small objects. Refine: eliminate such small objects. Bird bodies without necks and heads are good enough for us, for now.
connCompSize = table(connCompID)
connCompSizeconnCompID
0 1 2 3 4 5 6 7 8 9 10
477224 20 17 20 25 20 20 21 18 40 21
11 12 13 14 15 16 17 18 19 20 21
17 18 16 21 20 20 20 23 19 22 20
22 23 24 25 26 27 28 29 30 31 32
20 18 24 20 86 83 2 98 1 1 91
33 34 35 36 37 38 39 40 41 42 43
1 85 67 2 81 1 79 1 1 1 101
44 45 46 47 48 49 50 51 52 53 54
81 1 1 81 123 80 2 2 1 70 78
55 56 57 58 59 60 61 62 63 64 65
1 74 1 69 2 75 2 74 1 128 1
66 67 68 69 70 71
2 1 78 88 55 2 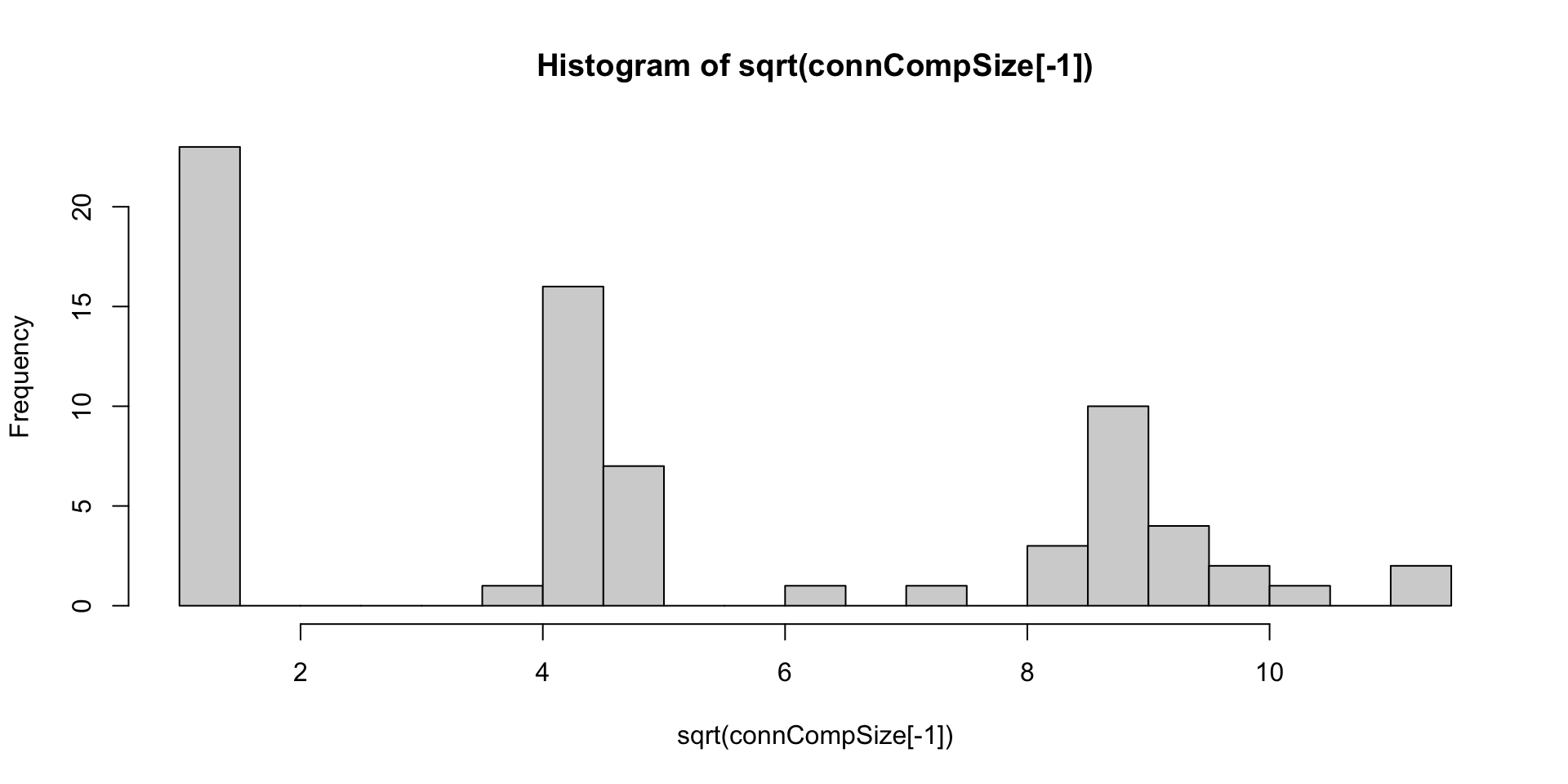
bad = names(connCompSize)[connCompSize < 4]
bad [1] "28" "30" "31" "33" "36" "38" "40" "41" "42" "45" "46" "50" "51" "52" "55"
[16] "57" "59" "61" "63" "65" "66" "67" "71"connCompID[ connCompID %in% as.integer(bad) ] = 0L
display(colorLabels(connCompID))
Count the number of birds. (Object “0” is the sky.)
Extract their coordinates (center of mass), and any other statistic we might care about.
library("dplyr")
birds = lapply(ids, function(i) {
w = which(connCompID == as.integer(i), arr.ind = TRUE)
tibble(
x = mean(w[, 1]),
y = mean(w[, 2]),
size = nrow(w),
phi = prcomp(w)$rotation[,1] |> (\(x) atan(x[1] / x[2]))())
}) |> bind_rows()
birds[1:3, ]# A tibble: 3 × 4
x y size phi
<dbl> <dbl> <int> <dbl>
1 559. 61.3 20 -0.138
2 550. 67.8 17 -0.208
3 526. 76.4 20 -0.166library("beyonce")
ggplot(birds, aes(x = x, y = -y, col = sqrt(size))) + geom_point() +
scale_color_gradientn(colors = beyonce_palette(72, 21, type = "continuous")) + coord_fixed()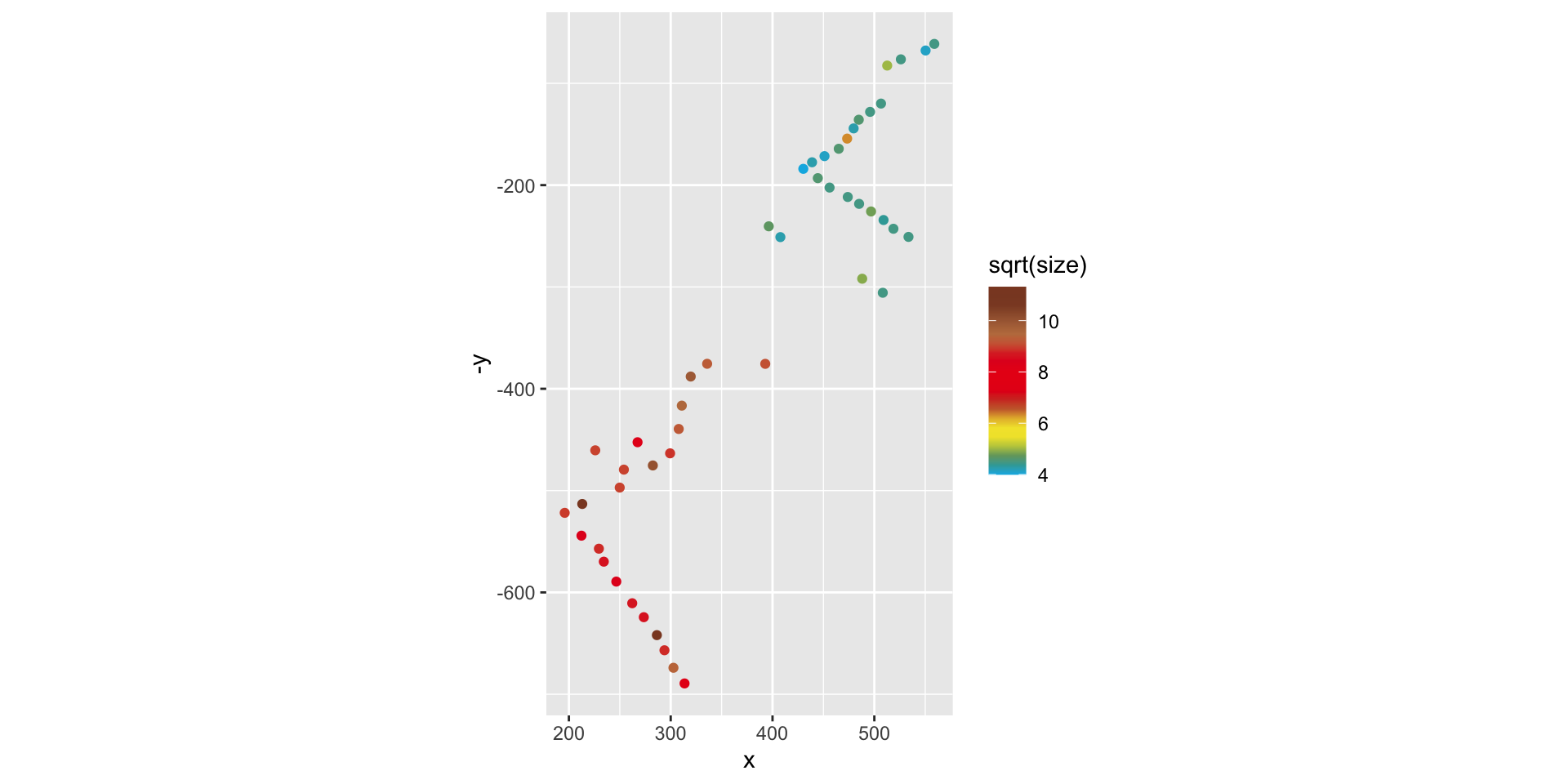
Optical flow analysis on a movie of bird murmuration
This video is by dylan.winter@virgin.net, and I got it via Youtube.
Use ffmpeg to extract the individual frames from time period 1:35 - 1:57 and store them as png files.
Use EBImage::readImages to read into 4D array: \(n_x\times n_y\times n_{\text{colors}}\times n_{\text{timepoints}}\).
Apply Optical Flow—basically, simple linear algebra / analysis—to detect and measure local velocities of image content
trsf = function(x, y) {
rv = atan2(y, x) / pi * 180
ifelse(rv <= (-175), rv+360, rv)
}
vec |>
dplyr::filter(abs(vx) + abs(vy) >= 2) |>
mutate(time = cut(t, 4),
angle = trsf(vx, vy)) |>
ggplot(aes(x = angle)) +
coord_polar(theta = "x", start = 95/180*pi, direction = -1) +
geom_histogram(binwidth = 15, center = 0) +
scale_x_continuous(breaks = seq(-180, 180, by = 15), expand = c(0, 0)) +
facet_wrap(vars(time), ncol = 2)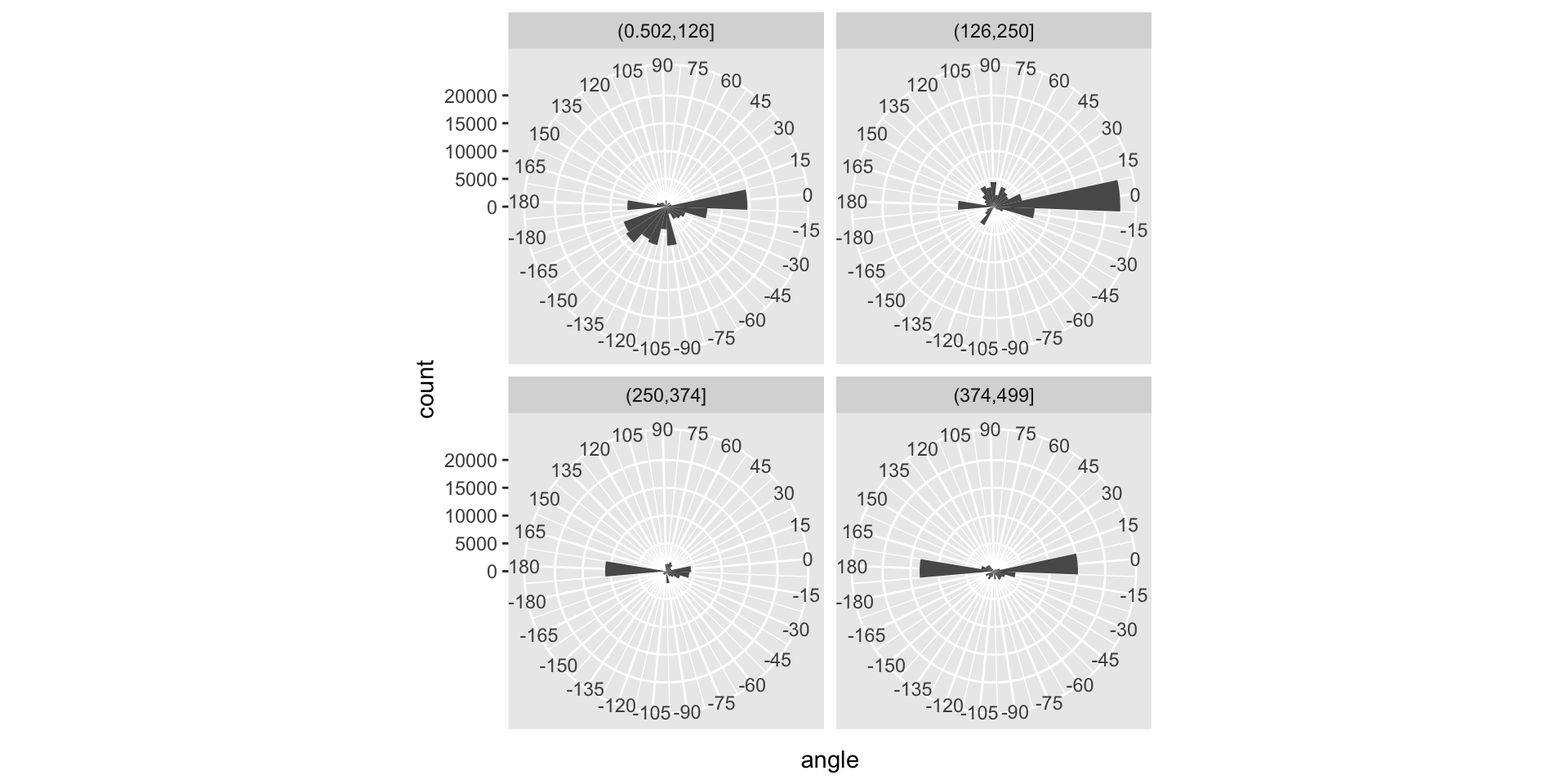
How does optical flow work?
display(grus) # time point 1
grus2 = translate(grus, c(30, 40))
display(grus2) # (simulated) time point 2

display(normalize(grus + grus2))
display(normalize(grus + translate(grus2, c(-5, 40))))
display(normalize(grus + translate(grus2, c(-30, -39))))


Try all possible translations of grus2 and find the one that leads to maximal overlap (correlation) with grus. imagefx::xcorr3d does this efficiently using FFT.
Full code for the bird murmuration example
# I first tried to download the video file with
youtube-dl "https://www.youtube.com/watch?v=eakKfY5aHmY"
# but this resulted in the error message also reported here https://stackoverflow.com/questions/75495800/error-unable-to-extract-uploader-id-youtube-discord-py
# So I followed the top-voted reply there, and ran
python3 -m pip install --force-reinstall https://github.com/yt-dlp/yt-dlp/archive/master.tar.gz
yt-dlp "https://www.youtube.com/watch?v=eakKfY5aHmY"
# The video has 25 frames per second. Some of the interesting segments are: 0:18-0:31, 1:21-1:33, 1:34-1:57, 2:10-2:32, 3:34-3:46
# I used the following to extract the frames from time period 1:35 - 1:57.
ffmpeg -ss 00:01:35 -t 00:01:57 -i resources/eakKfY5aHmY.mp4 frames/murm-%04d.pngRead the frames (png files) produced by ffmpeg
frames = dir("frames", full.names = TRUE)
frames = frames[1:500]
mov = readImage(frames)
print(object.size(mov), unit = "Gb")
movg = mov[,,1,] + mov[,,2,] + mov[,,3,]
colorMode(movg) = "grayscale"Optical flow analysis: manually divide the image into overlapping squares on a grid, centered around cx, cy, of side length 2*epsilon. Within each of them, for each time point, compute the flow vector fvec.
stride = 30
epsilon = 40
time = 1:dim(mov)[4]
# Instead of the 3 nested loops and fvec array, could also also use dplyr and a tidy tibble, depending on taste.
cx = seq(from = epsilon, to = dim(movg)[1] - epsilon, by = stride)
cy = seq(from = epsilon, to = dim(movg)[2] - epsilon, by = stride)
fvec = array(NA_real_, dim = c(4, length(cx), length(cy), length(time) - 1))
for(it in seq_len(length(time) - 1)) {
im1 = movg[, , time[it] ]
im2 = movg[, , time[it] + 1]
for(ix in seq(along = cx)) {
sx = (cx[ix] - epsilon + 1):(cx[ix] + epsilon)
for(iy in seq(along = cy)) {
sy = (cy[iy] - epsilon + 1):(cy[iy] + epsilon)
xc = imagefx::xcorr3d(im1[ sx, sy], im2[ sx, sy])
fvec[, ix, iy, it] = with(xc, c(max.shifts, max.corr, corr.mat[nrow(corr.mat)/2+1, ncol(corr.mat)/2+1]))
}
}
}
save(fvec, file = "resources/fvec.RData")Save each frame as a PNG.
scala = 3
for(it in seq_len(length(time) - 1)) {
png(file.path("opticalflow", sprintf("murm-%04d.png", it)), width = dim(mov)[1], height = dim(mov)[2], units = "px")
display(mov[,,,time[it]], method = "raster")
for(ix in seq(along = cx))
for(iy in seq(along = cy))
if (is.finite(fvec[3, ix, iy, it]) && (fvec[3, ix, iy, it] > 0.55) && any(fvec[1:2, ix, iy, it] != 0))
arrows(x0 = cx[ix], x1 = cx[ix] + scala * fvec[1, ix, iy, it],
y0 = cy[iy], y1 = cy[iy] + scala * fvec[2, ix, iy, it],
col = "#FFDD00", lwd = 2, length = 0.04)
dev.off()
}Assemble into a movie using ffmpeg.
RBioFormats
R interface to the Bio-Formats library by the Open Microscopy Environment (OME) collaboration for reading and writing image data in many different formats, incl. proprietary (vendor-specific) microscopy image data and metadata files.
Zarr and Rarr
The Zarr specification defines a format for chunked, compressed, N-dimensional arrays. It’s design allows efficient access to subsets of the stored array, and supports both local and cloud storage systems. Zarr is experiencing increasing adoption in a number of scientific fields, where multi-dimensional data are prevalent.
vitessce
session_info
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.2.3 (2023-03-15)
os macOS Ventura 13.2.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Berlin
date 2023-03-23
pandoc 2.19.2 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.2.0)
beyonce * 0.1 2023-03-18 [1] Github (dill/beyonce@d0a5316)
BiocGenerics 0.44.0 2022-11-07 [1] Bioconductor
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.2.0)
cachem 1.0.7 2023-02-24 [1] CRAN (R 4.2.0)
callr 3.7.3 2022-11-02 [1] CRAN (R 4.2.0)
cli 3.6.0 2023-01-09 [1] CRAN (R 4.2.0)
codetools 0.2-19 2023-02-01 [1] CRAN (R 4.2.3)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.2.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.2.0)
devtools 2.4.5 2022-10-11 [1] CRAN (R 4.2.0)
digest 0.6.31 2022-12-11 [1] CRAN (R 4.2.0)
dplyr * 1.1.0 2023-01-29 [1] CRAN (R 4.2.0)
EBImage * 4.40.0 2022-11-07 [1] Bioconductor
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.2.0)
evaluate 0.20 2023-01-17 [1] CRAN (R 4.2.0)
fansi 1.0.4 2023-01-22 [1] CRAN (R 4.2.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.2.0)
fftwtools 0.9-11 2021-03-01 [1] CRAN (R 4.2.0)
fs 1.6.1 2023-02-06 [1] CRAN (R 4.2.0)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.0)
ggplot2 * 3.4.1 2023-02-10 [1] CRAN (R 4.2.0)
glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
gtable 0.3.2 2023-03-17 [1] CRAN (R 4.2.0)
htmltools 0.5.4 2022-12-07 [1] CRAN (R 4.2.0)
htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.2.0)
httpuv 1.6.9 2023-02-14 [1] CRAN (R 4.2.0)
jpeg 0.1-10 2022-11-29 [1] CRAN (R 4.2.0)
jsonlite 1.8.4 2022-12-06 [1] CRAN (R 4.2.0)
knitr 1.42 2023-01-25 [1] CRAN (R 4.2.0)
later 1.3.0 2021-08-18 [1] CRAN (R 4.2.0)
lattice 0.20-45 2021-09-22 [1] CRAN (R 4.2.3)
lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.0)
locfit 1.5-9.7 2023-01-02 [1] CRAN (R 4.2.0)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
mime 0.12 2021-09-28 [1] CRAN (R 4.2.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.2.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.0)
pkgbuild 1.4.0 2022-11-27 [1] CRAN (R 4.2.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
pkgload 1.3.2 2022-11-16 [1] CRAN (R 4.2.0)
plyr 1.8.8 2022-11-11 [1] CRAN (R 4.2.0)
png 0.1-8 2022-11-29 [1] CRAN (R 4.2.0)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.2.0)
processx 3.8.0 2022-10-26 [1] CRAN (R 4.2.0)
profvis 0.3.7 2020-11-02 [1] CRAN (R 4.2.0)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.2.0)
ps 1.7.2 2022-10-26 [1] CRAN (R 4.2.0)
purrr 1.0.1 2023-01-10 [1] CRAN (R 4.2.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.2.0)
Rcpp 1.0.10 2023-01-22 [1] CRAN (R 4.2.0)
RCurl 1.98-1.10 2023-01-27 [1] CRAN (R 4.2.0)
remotes 2.4.2 2021-11-30 [1] CRAN (R 4.2.0)
reshape2 * 1.4.4 2020-04-09 [1] CRAN (R 4.2.0)
rlang 1.1.0 2023-03-14 [1] CRAN (R 4.2.0)
rmarkdown 2.20 2023-01-19 [1] CRAN (R 4.2.0)
rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.0)
scales 1.2.1 2022-08-20 [1] CRAN (R 4.2.0)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
shiny 1.7.4 2022-12-15 [1] CRAN (R 4.2.0)
stringi 1.7.12 2023-01-11 [1] CRAN (R 4.2.0)
stringr 1.5.0 2022-12-02 [1] CRAN (R 4.2.0)
terrainr * 0.7.4 2023-02-16 [1] CRAN (R 4.2.0)
tibble 3.2.1 2023-03-20 [1] CRAN (R 4.2.0)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.0)
tiff 0.1-11 2022-01-31 [1] CRAN (R 4.2.0)
unifir 0.2.3 2022-12-02 [1] CRAN (R 4.2.0)
urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.2.0)
usethis 2.1.6 2022-05-25 [1] CRAN (R 4.2.0)
utf8 1.2.3 2023-01-31 [1] CRAN (R 4.2.0)
vctrs 0.6.0 2023-03-16 [1] CRAN (R 4.2.3)
withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
xfun 0.37 2023-01-31 [1] CRAN (R 4.2.0)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.2.0)
yaml 2.3.7 2023-01-23 [1] CRAN (R 4.2.0)
[1] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────









