Identify drug responses associated with genomics in CLL
Junyan Lu
12 January 2023
Last updated: 2023-01-12
Checks: 6 1
Knit directory: EMBL2016/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210512) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 12d1722. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/CDK_analysis_cache/
Ignored: analysis/boxplot_AUC.png
Ignored: analysis/consensus_clustering_CPS_cache/
Ignored: analysis/consensus_clustering_noFit_cache/
Ignored: analysis/dose_curve.png
Ignored: analysis/targetDist.png
Ignored: analysis/toxivity_box.png
Ignored: analysis/volcano.png
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/AUC_CLL_IC50/
Untracked: analysis/BRAF_analysis.Rmd
Untracked: analysis/CDK_analysis.Rmd
Untracked: analysis/GSVA_analysis.Rmd
Untracked: analysis/MOFA_analysis.Rmd
Untracked: analysis/NOTCH1_signature.Rmd
Untracked: analysis/autoluminescence.Rmd
Untracked: analysis/bar_plot_mixed_noU1.pdf
Untracked: analysis/beatAML/
Untracked: analysis/consensus_clustering.Rmd
Untracked: analysis/consensus_clustering_CPS.Rmd
Untracked: analysis/consensus_clustering_IC50.Rmd
Untracked: analysis/consensus_clustering_beatAML.Rmd
Untracked: analysis/consensus_clustering_noFit.Rmd
Untracked: analysis/coxResTab.RData
Untracked: analysis/disease_specific.Rmd
Untracked: analysis/drugScreens_reproducibility.Rmd
Untracked: analysis/genomic_association.Rmd
Untracked: analysis/genomic_association_IC50.Rmd
Untracked: analysis/genomic_association_allDisease.Rmd
Untracked: analysis/noFit_CLL/
Untracked: analysis/outcome_associations.Rmd
Untracked: analysis/overview.Rmd
Untracked: analysis/plotCohort.Rmd
Untracked: analysis/preprocess.Rmd
Untracked: analysis/volcano_drugGene.pdf
Untracked: code/utils.R
Untracked: data/BeatAML_Waves1_2/
Untracked: data/ic50Tab.RData
Untracked: data/newEMBL_20210806.RData
Untracked: data/patMeta.RData
Untracked: data/targetAnnotation_all.csv
Untracked: output/gene_associations/
Untracked: output/mofaRes.rds
Untracked: output/resConsClust.RData
Untracked: output/resConsClust_aucFit.RData
Untracked: output/resConsClust_beatAML.RData
Untracked: output/resConsClust_cps.RData
Untracked: output/resConsClust_ic50.RData
Untracked: output/resConsClust_noFit.RData
Untracked: output/screenData.RData
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/_site.yml
Deleted: analysis/about.Rmd
Modified: analysis/index.Rmd
Deleted: analysis/license.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Load libraries and dataset
Load datasets
Overview of associations between drug responses and genomics in CLL
Using AUC of trepazoidal rule
Without blocking for IGHV
Only mutations occcured at least 5 times will be included in the test
Visualize the mean effect and variance of each drug
meanSdTab <- tibble(name = rownames(viabMat),
meanVal = rowMeans(viabMat, na.rm = TRUE),
sdVal = genefilter::rowSds(viabMat, na.rm=TRUE))
ggplot(meanSdTab, aes(x=meanVal, y=sdVal)) + geom_point()
Some drugs perhaps don’t have any effect at all and including them may increase multiple hypothesis testing burden, resulting less associations pass 10% FDR. There are perhaps two ways to deal with this: 1) prefiltering the drugs or using IHW and mean effect + sd as covariate.
Perform test
Adjust p-value use IHW, using standard deviation as covariate
ihwTab <- tibble(pval = pTab$p, name = pTab$drug) %>%
left_join(meanSdTab)
ihwRes <- ihw(pval ~ sdVal, data = ihwTab, alpha = 0.1)
pTab$p.adj.ihw <- adj_pvalues(ihwRes)
#plot(ihwRes)Write out test result table
write_csv2(pTab,"../docs/p_table_noBlock.csv")Number of significant associations per gene (10% FDR)

Number of significant associations per gene (10% FDR), adjusted by IHW
 Seems to help a little, especially with some each associations.
Seems to help a little, especially with some each associations.
Perform test after pre-filtering non-effective drugs
#keepDrug <- filter(meanSdTab, meanVal < 0.9, sdVal > 0.05)$name #a rather arbitrary cutoff
viabMat.filt <- viabMat#[keepDrug, ]
pTab.filt <- lapply(colnames(geneBack), function(geneName) {
var <- geneBack[,geneName, drop=FALSE]
designMat <- model.matrix(~., data = var)
testMat <- viabMat.filt[,rownames(designMat)]
fit <- lmFit(testMat, designMat)
fit2 <- eBayes(fit)
res <- topTable(fit2, number = "all") %>%
as_tibble(rownames = "drug") %>%
mutate(gene = geneName) %>%
select(drug, gene, P.Value, adj.P.Val, logFC, t) %>%
dplyr::rename(p=P.Value, p.adj = adj.P.Val)
}) %>% bind_rows() %>%
left_join(select(targetAnno, drugName, target, pathway, targetFamily), by = c( drug = "drugName"))
plotTab <- filter(pTab.filt, p.adj <= 0.1) %>% group_by(gene) %>%
summarise(number = length(drug)) %>%
arrange(desc(number)) %>% mutate(gene = factor(gene, levels = gene))
ggplot(plotTab, aes(x=gene, y = number, fill = gene)) +
geom_bar(stat = "identity", show.legend = FALSE) +
geom_text(aes(label=number), position=position_dodge(width=0.9), vjust=-0.25) +
theme_bw() + ggplot2::theme(axis.text.x = element_text(angle = 90, hjust =1, vjust = .5)) +
xlab("Variant name") + ylab("Number of significant associations (10% FDR)")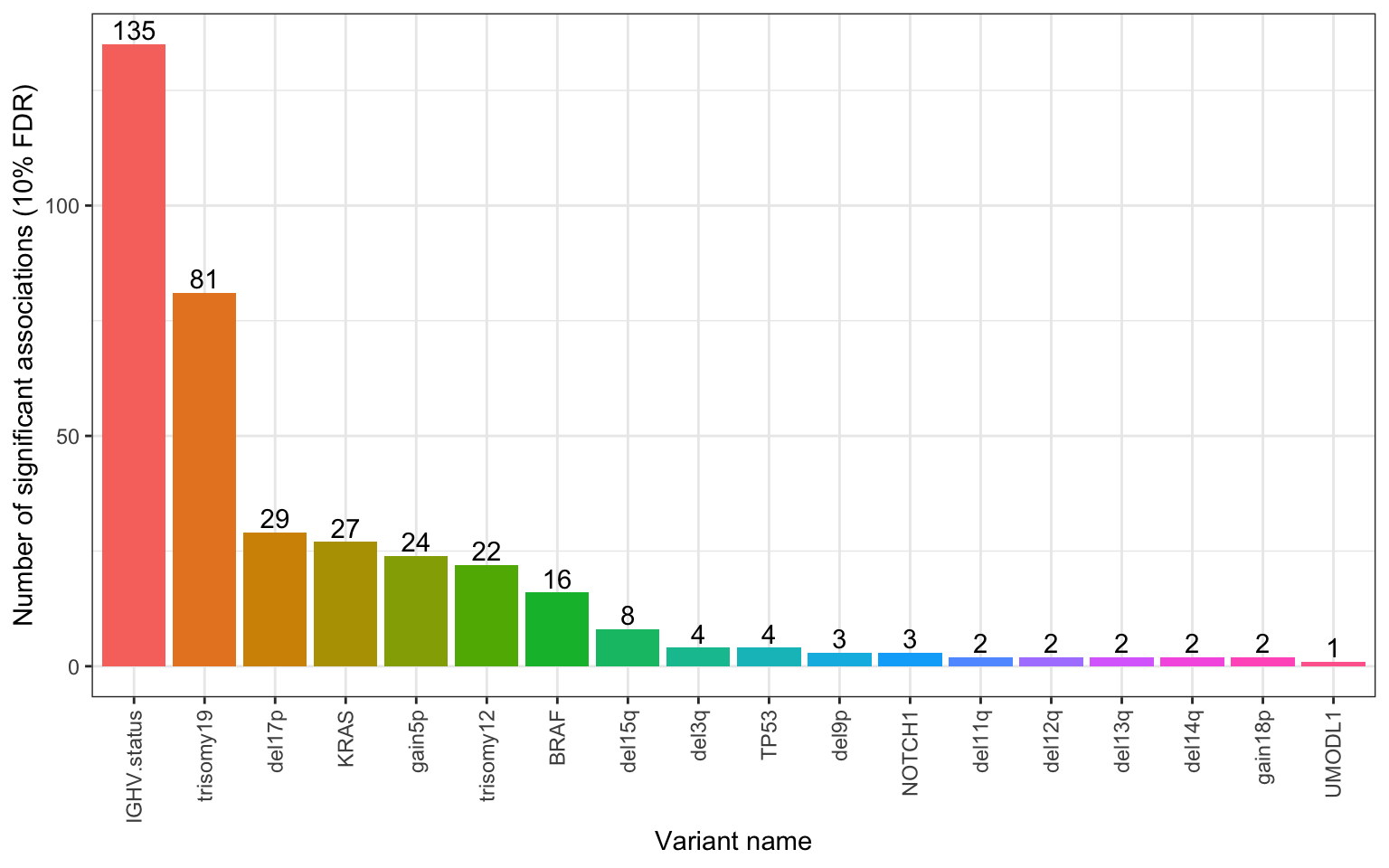 May also help a little.
May also help a little.
Without IHW
#pTab <- mutate(pTab, p.adj = p.adj.ihw)P value scatter plot (only show drugs with assocations)
 Associations pass 10% FDR are colored by genes.
Associations pass 10% FDR are colored by genes.
Only a few associations pass the 10% FDR threshold, although many
associations pass raw p-value 0.01 threshold. This could be due to the
multiple hypothesis testing problem. We have more drugs in EMLB2016
screen than other screens. (I already pre-filtered the drugs that show
very little variance across samples.)
PDF version: pScatter-1.pdf
Volcano plots (per gene)
#filter genes with significant assocaitions
useGene <- unique(filter(pTab, p.adj <=0.1)$gene)
#get top 10 most up and down regulated genes
upDrug <- lapply(unique(pTab$gene), function(n) {
dplyr::filter(pTab, gene ==n, logFC >0) %>% top_n(10, -log10(p))
}) %>% bind_rows()
downDrug <- lapply(unique(pTab$gene), function(n) {
dplyr::filter(pTab, gene == n, logFC < 0) %>% top_n(10, -log10(p))
}) %>% bind_rows()
drugLab <- bind_rows(upDrug, downDrug) %>%
filter(p.adj <0.1) %>%
mutate(drugLabel = drug) %>% select(drug, gene, drugLabel)
plotList <- lapply(useGene, function(n) {
eachTab <- filter(pTab, gene %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
left_join(drugLab, by = c("drug", "gene"))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- -log10(0.1)
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(aes(label = drugLabel), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s (mutated vs unmutated)", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_noBlocking.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_noBlocking.pdf
Volcano plots (per drug)
Only drugs with FDR < 0.1 with at least one gene
#filter genes with significant assocaitions
useDrug <- unique(filter(pTab, p.adj <= 0.1)$drug)
plotList <- lapply(useDrug, function(n) {
eachTab <- filter(pTab, drug %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive")))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- 1
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(data = filter(eachTab, direction != "n.s."), aes(label = gene), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
#plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_perDrug_noBlocking.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_perDrug_noBlocking.pdf
Volcano plots (drug+gene)
mutNum <- colSums(geneBack,na.rm=TRUE)
plotTab <- pTab %>% mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(num = mutNum[gene]) %>%
mutate(featureName = paste0(drug,"\n",gene))
labelFeature <- arrange(plotTab, t)$featureName[c(1:10, (nrow(plotTab)-9):nrow(plotTab))]
plotTab <- mutate(plotTab, ifLabel = featureName %in% labelFeature)
pCut <- 1
ggplot(plotTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(fill = direction, size = num), shape = 21, alpha=0.5) +
ggrepel::geom_text_repel(data = filter(plotTab, ifLabel),
aes(label = featureName),
max.overlaps = 100) +
scale_fill_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
ggsave("../docs/volcano_drugGene.pdf", height = 10, width = 10)Only top 10 most associations in each direction are labeled pdf file
Boxplots and dose-response curves per gene
pTab.sig <- filter(pTab, p.adj < 0.1)
if (!dir.exists("../output/gene_associations")) dir.create("../output/gene_associations")
plotList <- lapply(unique(pTab.sig$gene), function(n) {
eachTab <- filter(pTab.sig, gene == n)
eachGenePlot <- lapply(seq(nrow(eachTab)), function(i) {
rec <- eachTab[i,]
plotTab <- filter(screenData, Drug == rec$drug, diagnosis =="CLL") %>%
mutate(mut = factor(geneBack[match(patientID,rownames(geneBack)),][[rec$gene]])) %>%
filter(!is.na(mut)) %>%
group_by(patientID, conc, mut) %>%
summarise(viab=mean(viab, na.rm=TRUE), viab.auc = mean(viab.auc,na.rm=TRUE)) %>%
ungroup() %>% arrange(mut) %>% mutate(patientID = factor(patientID, levels = unique(patientID)))
plotTab.auc <- distinct(plotTab, patientID, mut, viab.auc)
pBox <- ggplot(plotTab.auc, aes(x=mut, y=viab.auc)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(aes(col = mut)) +
theme_bw() +ylab("Viability") + xlab("mutation status") +
ggtitle(sprintf("%s (p.adj=%s)",rec$drug,formatC(rec$p.adj, digits = 2))) +
theme(legend.position = "none")
pDose <- ggplot(plotTab, aes(x=conc, y=viab, col = mut, group = patientID)) +
geom_point() + geom_smooth(method="loess",se=FALSE, formula = y~x) +
scale_x_log10() +
theme_bw() +ylab("Viability") + xlab("concentration") +
ggtitle(sprintf("%s, %s",rec$pathway,rec$targetFamily))
pCom <- plot_grid(pBox, pDose, rel_widths = c(0.5,1))
pCom
})
jyluMisc::makepdf(eachGenePlot , sprintf("../output/gene_associations/%s.pdf",n),ncol = 1,nrow = 4,width = 10, height = 12)
NULL
})Download link gene_associations.zip
A table of significant associations
filter(pTab, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
DT::datatable()Blocking for IGHV
Number of significant associations per gene (10% FDR)
 Associations pass 10% FDR are colored by genes.
Associations pass 10% FDR are colored by genes.
Number of significant associations per gene (10% FDR), adjusted by IHW
 IHW does not help too much here.
IHW does not help too much here.
Perform test after pre-filtering non-effective drugs
#keepDrug <- filter(meanSdTab, meanVal < 0.9, sdVal > 0.05)$name #a rather arbitrary cutoff
viabMat.filt <- viabMat#[keepDrug, ]
pTab.block.filt <- lapply(colnames(geneBack.noIGHV), function(geneName) {
var <- geneBack[,c("IGHV.status",geneName), drop=FALSE]
designMat <- model.matrix(~., data = var)
testMat <- viabMat.filt[,rownames(designMat)]
fit <- lmFit(testMat, designMat)
fit2 <- eBayes(fit)
res <- topTable(fit2, number = "all", coef = geneName) %>%
as_tibble(rownames = "drug") %>%
mutate(gene = geneName) %>%
select(drug, gene, P.Value, adj.P.Val, logFC, t) %>%
dplyr::rename(p=P.Value, p.adj = adj.P.Val)
}) %>% bind_rows() %>%
left_join(select(targetAnno, drugName, target, pathway, targetFamily), by = c( drug = "drugName"))
plotTab <- filter(pTab.block.filt, p.adj <= 0.1) %>% group_by(gene) %>%
summarise(number = length(drug)) %>%
arrange(desc(number)) %>% mutate(gene = factor(gene, levels = gene))
ggplot(plotTab, aes(x=gene, y = number, fill = gene)) +
geom_bar(stat = "identity", show.legend = FALSE) +
geom_text(aes(label=number), position=position_dodge(width=0.9), vjust=-0.25) +
theme_bw() + ggplot2::theme(axis.text.x = element_text(angle = 90, hjust =1, vjust = .5)) +
xlab("Variant name") + ylab("Number of significant associations (10% FDR)")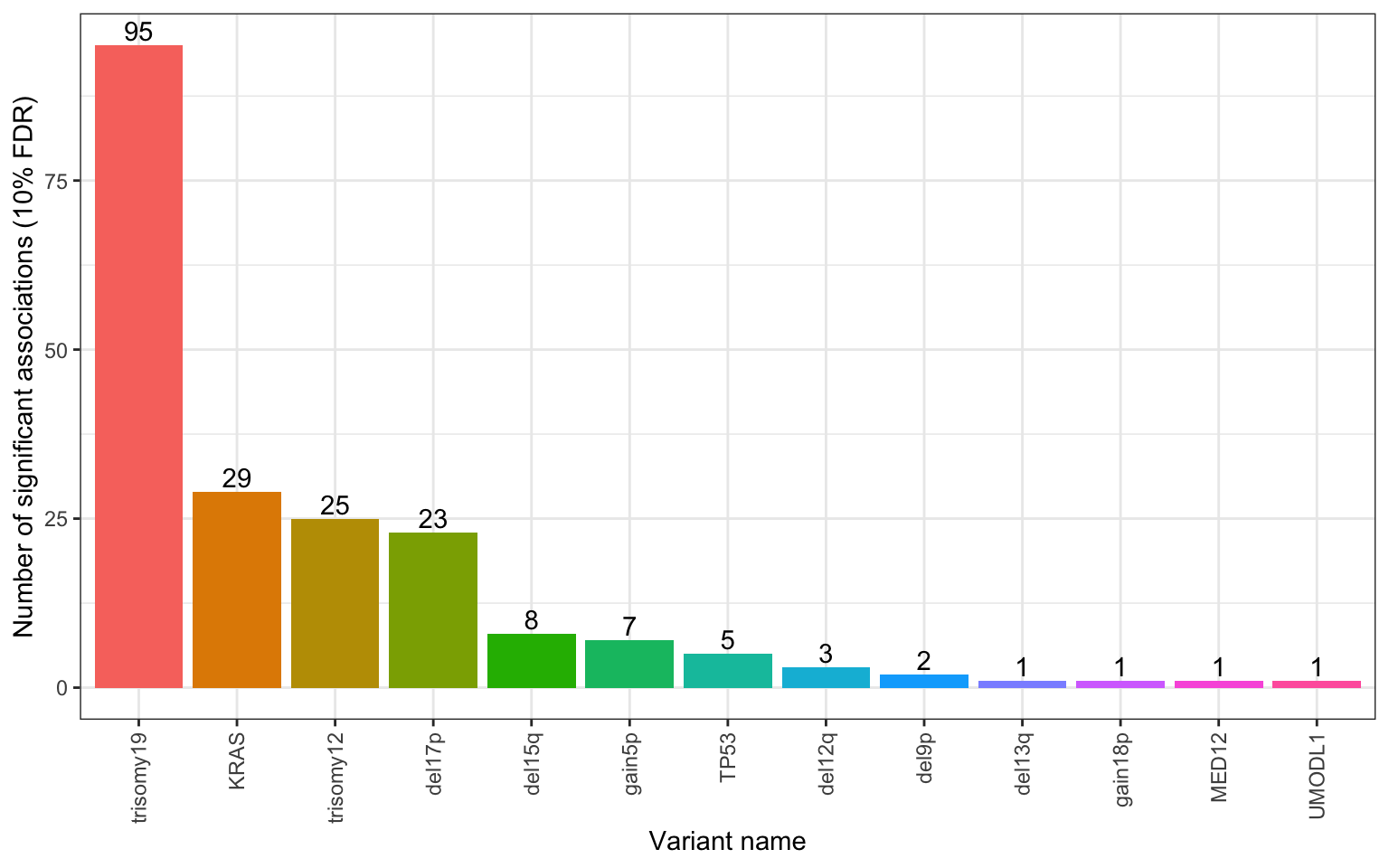 Pre-filtering actually is better here.
Pre-filtering actually is better here.
Number of significant associations per gene, blocking for non-IGHV features

Number of significant associations per gene, blocking for non-IGHV features, without U1

P value scatter plot (with pre-filtering)
pTab.block <- pTab.block.filt PDF version: pScatter_aov-1.pdf
PDF version: pScatter_aov-1.pdf
Volcano plots
#filter genes with significant assocaitions
useGene <- unique(filter(pTab.block, p.adj <=0.1)$gene)
#get top 10 most up and down regulated genes
upDrug <- lapply(unique(pTab.block$gene), function(n) {
filter(pTab.block, gene ==n, logFC >0) %>% top_n(10, -log10(p))
}) %>% bind_rows()
downDrug <- lapply(unique(pTab.block$gene), function(n) {
filter(pTab.block, gene == n, logFC < 0) %>% top_n(10, -log10(p))
}) %>% bind_rows()
drugLab <- bind_rows(upDrug, downDrug) %>%
filter(p.adj <0.1) %>%
mutate(drugLabel = drug) %>% select(drug, gene, drugLabel)
plotList <- lapply(useGene, function(n) {
eachTab <- filter(pTab.block, gene %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
left_join(drugLab, by = c("drug", "gene"))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- -log10(0.1)
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(aes(label = drugLabel), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s (mutated vs unmutated)", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_withBlocking.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_withBlocking.pdf
Volcano plots (per drug)
Only drugs with FDR < 0.1 with at least one gene
#filter genes with significant assocaitions
useDrug <- unique(filter(pTab.block, p.adj <= 0.1)$drug)
plotList <- lapply(useDrug, function(n) {
eachTab <- filter(pTab.block, drug %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive")))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- 1
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(data = filter(eachTab, direction != "n.s."), aes(label = gene), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
#plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_perDrug_withBlocking.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_perDrug_withBlocking.pdf
Volcano plots (drug+gene)
mutNum <- colSums(geneBack,na.rm=TRUE)
plotTab <- pTab.block %>% mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(num = mutNum[gene]) %>%
mutate(featureName = paste0(drug,"\n",gene))
labelFeature <- arrange(plotTab, t)$featureName[c(1:10, (nrow(plotTab)-9):nrow(plotTab))]
plotTab <- mutate(plotTab, ifLabel = featureName %in% labelFeature)
pCut <- 1
ggplot(plotTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(fill = direction, size = num), shape = 21, alpha=0.5) +
ggrepel::geom_text_repel(data = filter(plotTab, ifLabel),
aes(label = featureName),
max.overlaps = 100) +
scale_fill_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))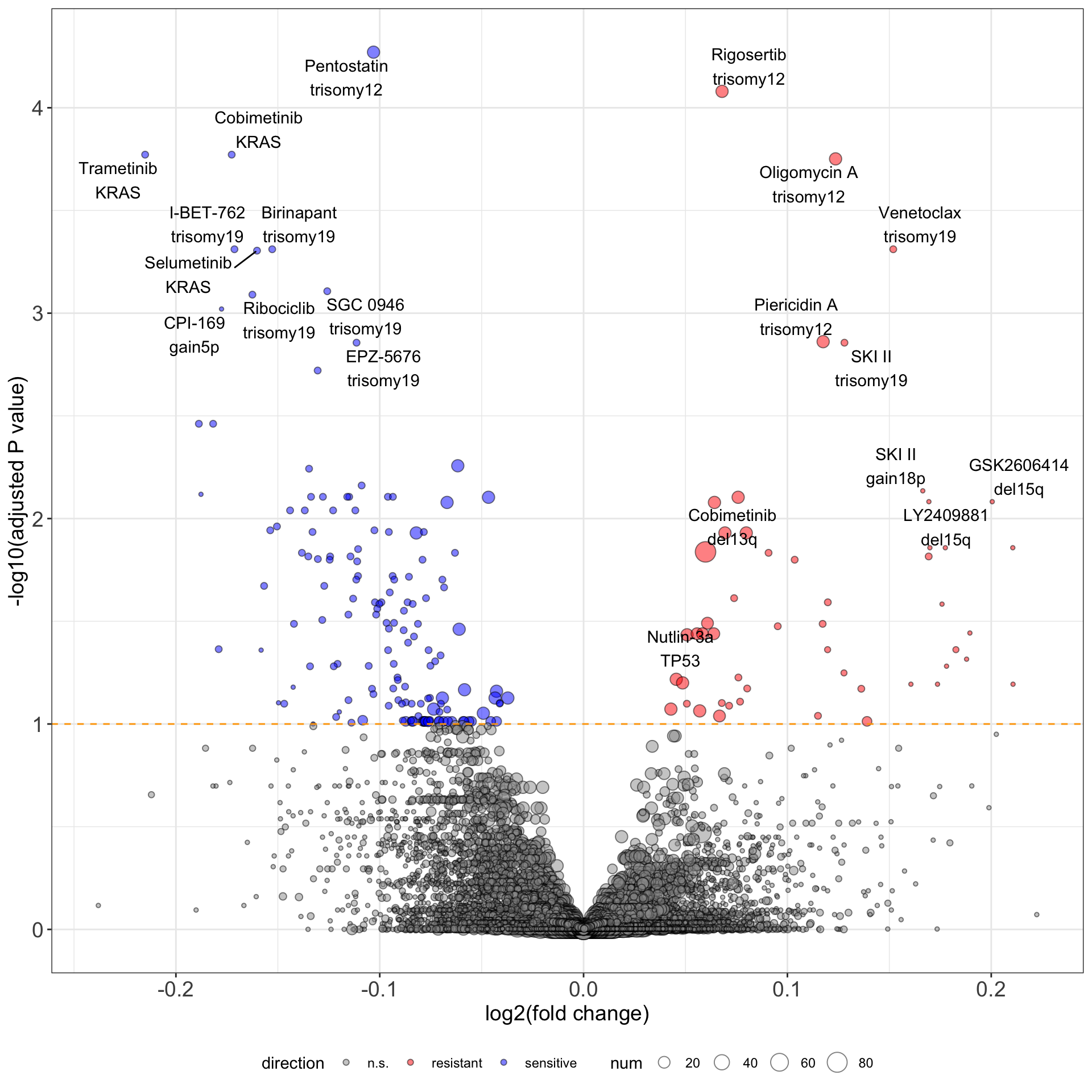
ggsave("../docs/volcano_drugGene_IGHVblocked.pdf")Only top 10 most associations in each direction are labeled pdf file
A table of significant associations
filter(pTab.block, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
DT::datatable()Within M-CLL only
Only mutations occcured at least 3 times will be included in the test
Number of significant associations per gene (10% FDR)

Volcano plots
#filter genes with significant assocaitions
useGene <- unique(filter(pTab, p.adj <=0.1)$gene)
plotList <- lapply(useGene, function(n) {
eachTab <- filter(pTab, gene %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(drugLabel = ifelse(direction == "n.s.","",drug))
pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
ggplot(eachTab, aes(x=logFC, y = -log10(p))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(aes(label = drugLabel), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10 (P value)") + xlab("log2 Fold Change") +
ggtitle(sprintf("%s (mutated vs unmutated)", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom")
})
plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_M_CLL.pdf", ncol=1, nrow=1, height = 12, width = 12)PDF version: volcano_M_CLL.pdf
Volcano plots (per drug)
Only drugs with FDR < 0.1 with at least one gene
#filter genes with significant assocaitions
useDrug <- unique(filter(pTab, p.adj <= 0.1)$drug)
plotList <- lapply(useDrug, function(n) {
eachTab <- filter(pTab, drug %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive")))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- 1
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(data = filter(eachTab, direction != "n.s."), aes(label = gene), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
#plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_perDrug_M_CLL.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_perDrug_M_CLL.pdf
Volcano plots (drug+gene)
mutNum <- colSums(geneBack,na.rm=TRUE)
plotTab <- pTab %>% mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(num = mutNum[gene]) %>%
mutate(featureName = paste0(drug,"\n",gene))
labelFeature <- arrange(plotTab, t)$featureName[c(1:10, (nrow(plotTab)-9):nrow(plotTab))]
plotTab <- mutate(plotTab, ifLabel = featureName %in% labelFeature)
pCut <- 1
ggplot(plotTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(fill = direction, size = num), shape = 21, alpha=0.5) +
ggrepel::geom_text_repel(data = filter(plotTab, ifLabel),
aes(label = featureName),
max.overlaps = 100) +
scale_fill_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
ggsave("../docs/volcano_drugGene_M.pdf", height = 10, width = 10)Only top 10 most associations in each direction are labeled pdf file
A table of significant associations
filter(pTab, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
DT::datatable()Within U-CLL only
Only mutations occcured at least 3 times will be included in the test
Number of significant associations per gene (10% FDR)

Volcano plots
#filter genes with significant assocaitions
useGene <- unique(filter(pTab, p.adj <=0.1)$gene)
plotList <- lapply(useGene, function(n) {
eachTab <- filter(pTab, gene %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(drugLabel = ifelse(direction == "n.s.","",drug))
pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
ggplot(eachTab, aes(x=logFC, y = -log10(p))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(aes(label = drugLabel), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10 (P value)") + xlab("log2 Fold Change") +
ggtitle(sprintf("%s (mutated vs unmutated)", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom")
})
plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_U_CLL.pdf", ncol=1, nrow=1, height = 12, width = 12)PDF version: volcano_U_CLL.pdf
Volcano plots (per drug)
Only drugs with FDR < 0.1 with at least one gene
#filter genes with significant assocaitions
useDrug <- unique(filter(pTab, p.adj <= 0.1)$drug)
plotList <- lapply(useDrug, function(n) {
eachTab <- filter(pTab, drug %in% n, !is.na(p)) %>%
mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive")))
#pCut <- -log10(max(filter(eachTab, p.adj <=0.1)$p))
pCut <- 1
ggplot(eachTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(col = direction)) +
ggrepel::geom_text_repel(data = filter(eachTab, direction != "n.s."), aes(label = gene), max.overlaps = 100) +
scale_color_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
ggtitle(sprintf("%s", n)) +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
})
#plot_grid(plotlist = plotList, ncol=2)
makepdf(plotList, "../docs/volcano_perDrug_U_CLL.pdf", ncol=2, nrow=2, height = 10, width = 9)PDF version: volcano_perDrug_U_CLL.pdf
Volcano plots (drug+gene)
mutNum <- colSums(geneBack,na.rm=TRUE)
plotTab <- pTab %>% mutate(direction = ifelse(p.adj > 0.1, "n.s.",
ifelse(logFC>0, "resistant","sensitive"))) %>%
mutate(num = mutNum[gene]) %>%
mutate(featureName = paste0(drug,"\n",gene))
labelFeature <- arrange(plotTab, t)$featureName[c(1:10, (nrow(plotTab)-9):nrow(plotTab))]
plotTab <- mutate(plotTab, ifLabel = featureName %in% labelFeature)
pCut <- 1
ggplot(plotTab, aes(x=logFC, y = -log10(p.adj))) +
geom_point(aes(fill = direction, size = num), shape = 21, alpha=0.5) +
ggrepel::geom_text_repel(data = filter(plotTab, ifLabel),
aes(label = featureName),
max.overlaps = 100) +
scale_fill_manual(values = c("n.s."="grey60","resistant" = "red","sensitive" = "blue")) +
geom_hline(yintercept = pCut, linetype = "dashed", color = "orange") +
ylab("-log10(adjusted P value)") + xlab("log2(fold change)") +
theme_bw() +
theme(plot.title = element_text(hjust=0.5, size=15, face ="bold"),
legend.position = "bottom",
axis.text = element_text(size=14),
axis.title = element_text(size=14))
ggsave("../docs/volcano_drugGene_U.pdf", height = 10, width = 10)Only top 10 most associations in each direction are labeled pdf file
A table of significant associations
filter(pTab, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
DT::datatable()Using individual concentrations
Without blocking for IGHV
Number of significant associations per gene (10%
FDR)

P value heatmap
Only drugs show at least one significant association under 10% FDR
pTab.sig <- filter(pTab, p.adj <= 0.1)
plotTab <- filter(pTab, gene %in% pTab.sig$gene) %>%
filter(Drug %in% pTab.sig$Drug) %>%
mutate(sign = ifelse(p.adj <= 0.1, "*",""),
pSign = -log10(p)) %>%
mutate(pSign = ifelse(pSign > 12, 12, pSign)) %>%
mutate(pSign = pSign * sign(logFC),
Drug = sprintf("%s (%s)",Drug, targetFamily))
pMat <- mutate(plotTab, geneConc = paste0(gene,"_", concIndex)) %>%
select(Drug, geneConc, pSign) %>%
pivot_wider(names_from = geneConc, values_from = pSign) %>%
data.frame() %>% column_to_rownames("Drug")
hc <- hclust(dist(pMat))
drugOrder <- rownames(pMat)[hc$order]
plotTab <- mutate(plotTab, Drug = factor(Drug, levels = drugOrder),
gene = factor(gene, levels = levels(sumTab$gene)))
ggplot(plotTab, aes(x=concIndex, y = Drug, fill = pSign)) +
geom_tile() + geom_text(aes(label=sign), nudge_y = -0.25) +
scale_fill_gradient2(low = "blue", mid = "white", high = "red", name ="-log10(P-value)") +
facet_wrap(~gene, ncol =12) +
xlab("concentration index") * indicates assocations passed 10% FDR control
* indicates assocations passed 10% FDR control
A table of significant associations
filter(pTab, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
left_join(select(targetAnno, drugName, target, pathway), by = c(Drug = "drugName")) %>%
DT::datatable()Blocking for IGHV
Number of significant associations per gene (10%
FDR)
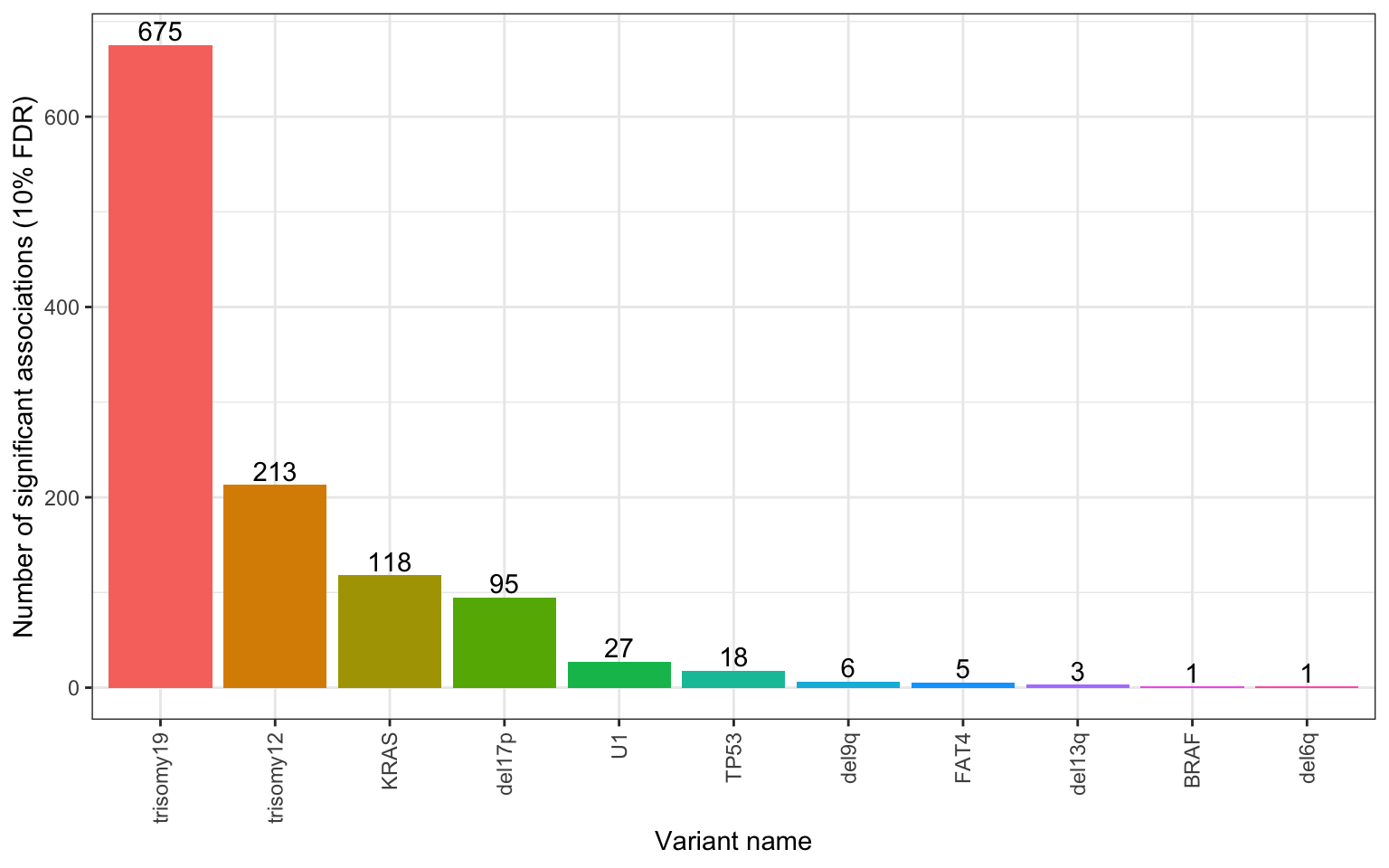
P value heatmap
Only drugs show at least one significant association under 10% FDR
pTab.sig <- filter(pTab, p.adj <= 0.1)
plotTab <- filter(pTab, gene %in% pTab.sig$gene) %>%
filter(Drug %in% pTab.sig$Drug) %>%
mutate(sign = ifelse(p.adj <= 0.1, "*",""),
pSign = -log10(p)) %>%
mutate(pSign = ifelse(pSign > 12, 12, pSign)) %>%
mutate(pSign = pSign * sign(logFC),
Drug = sprintf("%s (%s)",Drug, targetFamily))
pMat <- mutate(plotTab, geneConc = paste0(gene,"_", concIndex)) %>%
select(Drug, geneConc, pSign) %>%
pivot_wider(names_from = geneConc, values_from = pSign) %>%
data.frame() %>% column_to_rownames("Drug")
hc <- hclust(dist(pMat))
drugOrder <- rownames(pMat)[hc$order]
plotTab <- mutate(plotTab, Drug = factor(Drug, levels = drugOrder),
gene = factor(gene, levels = levels(sumTab$gene)))
ggplot(plotTab, aes(x=concIndex, y = Drug, fill = pSign)) +
geom_tile() + geom_text(aes(label=sign), nudge_y = -0.25) +
scale_fill_gradient2(low = "blue", mid = "white", high = "red", name = "-log10(P-value)") +
facet_wrap(~gene, ncol =12) +
xlab("concentration index") * indicates assocations passed 10% FDR control
* indicates assocations passed 10% FDR control
A table of significant associations
targetAnno <- read_csv2("../data/targetAnnotation_all.csv") %>%
mutate(drugName = nameEMBL2016)
filter(pTab, p.adj <=0.1) %>% mutate_if(is.numeric, formatC, digits=2) %>%
left_join(select(targetAnno, drugName, target, pathway), by = c(Drug = "drugName")) %>%
DT::datatable()Drug responses associated with IGHV
Volcano plot
 Drugs colored by blue are more effective in U-CLL samples. The names of
the drugs that show significant associations and effect size above 10%
in at least 3 concentrations are labeled. Dashed line indicates 5%
FDR
Drugs colored by blue are more effective in U-CLL samples. The names of
the drugs that show significant associations and effect size above 10%
in at least 3 concentrations are labeled. Dashed line indicates 5%
FDR
As expected, M-CLL samples show increased resistance to a lot of drugs.
Drug responses associated with Trisomy12
For all CLLs
How many triosmy12 samples?
tri12Tab <- distinct(viabTab, patientID, .keep_all = TRUE)
tri12Tab %>% filter(trisomy12 == 1) %>% nrow()[1] 23Volcano plot (10% FDR cut-off) for combined
concentrations
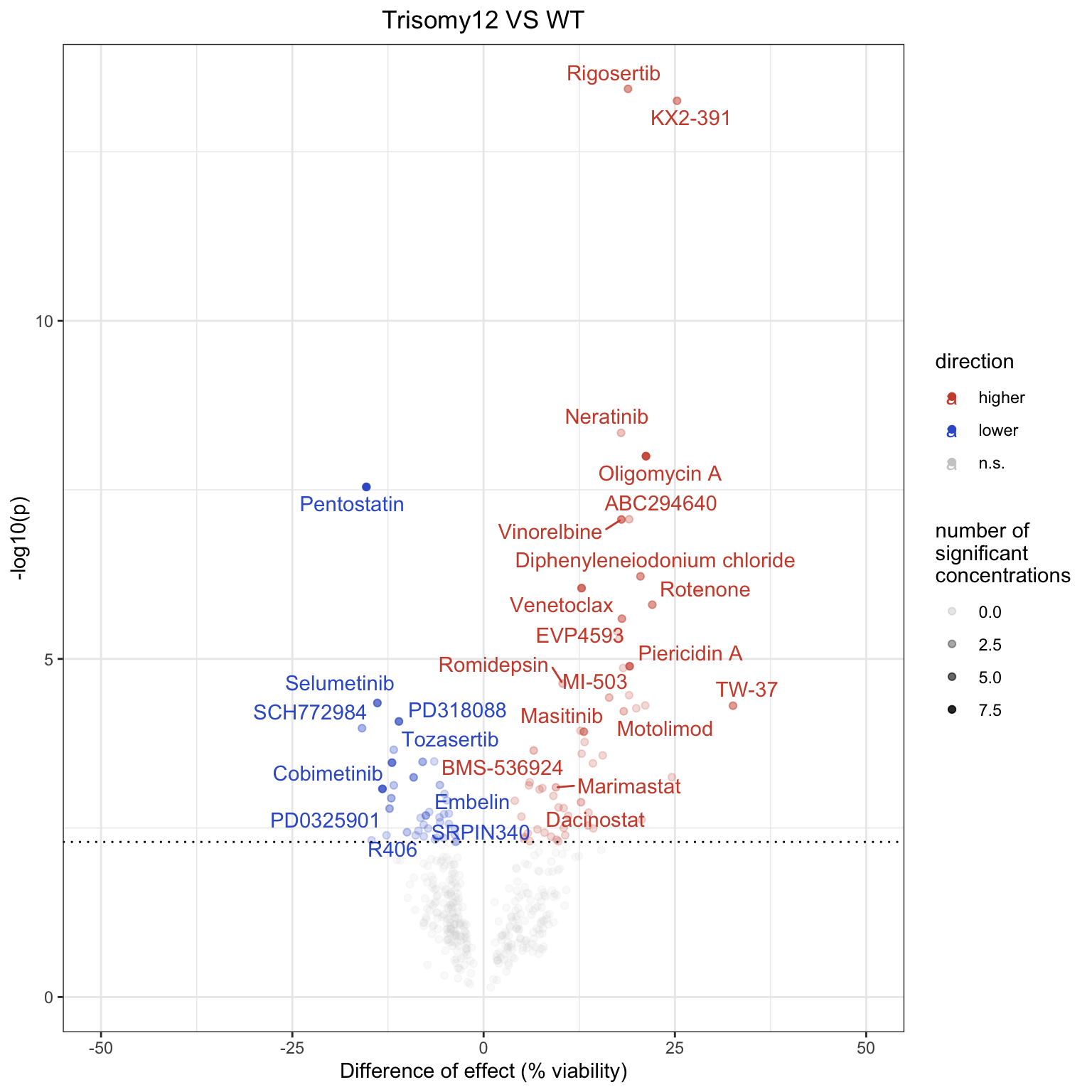 Drugs colored by blue are more effective in samples with trisomy12. The
names of the drugs that show significant associations in at least 2
concentrations are labeled. Dashed line indicates 10% FDR.
Drugs colored by blue are more effective in samples with trisomy12. The
names of the drugs that show significant associations in at least 2
concentrations are labeled. Dashed line indicates 10% FDR.
Compared to other datasets, more associations between increased drug resistance and trisomy12 are identified. But fewer associations between increased sensitivity and trisomy12 are identified.
Volcation plots for individual concentrations
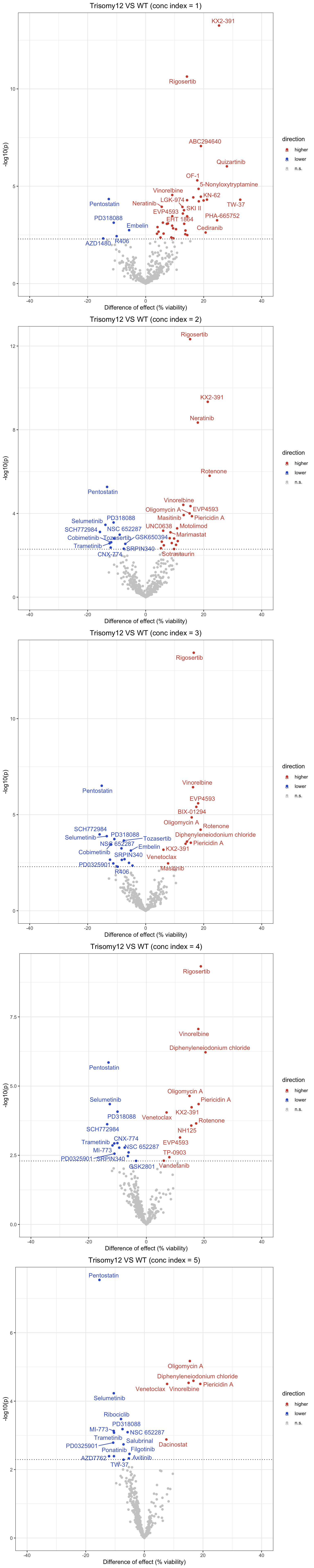
Beeswarm plots for all drug at all concentrations
For M CLLs only
Volcano plot (combined concentrations)

Volcation plots for individual concentrations
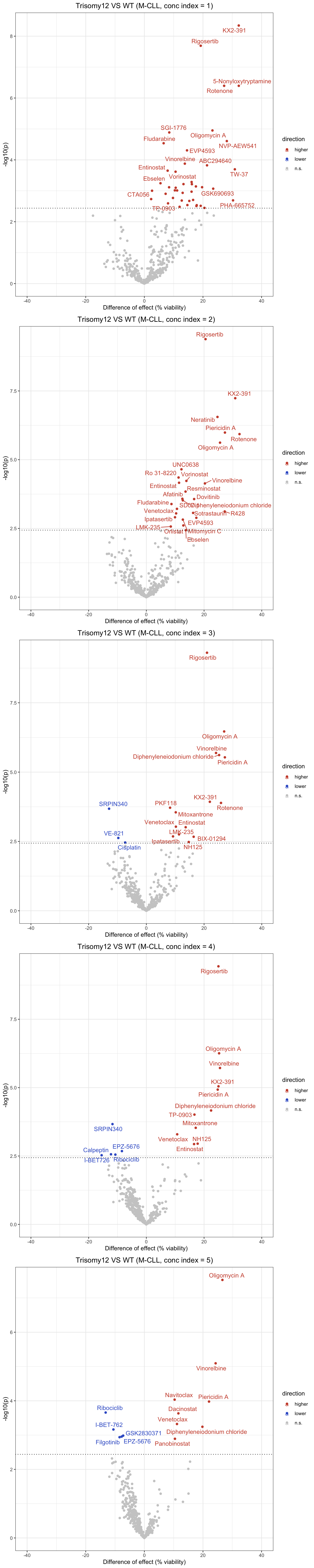
Beeswarm plots for all drug at all concentrations
For U CLLs
Volcano plot (combined concentrations)

Volcation plots for individual concentrations

Beeswarm plots for all drug at all concentrations
Drug responses associated with DDX3X
For U-CLLs only
viabTab <- screenData %>%
filter(diagnosis %in% "CLL") %>%
dplyr::select(patientID, viab, concIndex, Drug) %>%
group_by(patientID, concIndex, Drug) %>% summarise(viab= mean(viab)) %>%
ungroup() %>%
mutate(ighv = patBack[match(patientID, patBack$Patient.ID),]$IGHV.status,
ddx3x = patBack[match(patientID, patBack$Patient.ID),]$DDX3X)Correlation between DDX3X and IGHV
ddx3tab <- distinct(viabTab, patientID, .keep_all = TRUE)
table(ddx3tab$ddx3x, ddx3tab$ighv)
M U
0 67 55
1 0 5How many significant associations at 10% FDR?
filter(pTab, p.adj <= 0.1)# A tibble: 0 × 6
# … with 6 variables: Drug <fct>, concIndex <fct>, p <dbl>, diff <dbl>,
# p.adj <dbl>, ifSig <lgl>
# ℹ Use `colnames()` to see all variable namesNo significant associations, could be the DDX3X mutated cases are too few?
Volcano plot for combined concentrations (0.05 raw-palue
cut-off)

Volcation plots for individual concentrations
Note that the dash line indicates raw p value of 0.05, not
10% FDR

Beeswarm plots for all drug at all concentrations
Co-occurrence of genomic variance
Detecting interactions by chi-square test
chiTab <- lapply(seq(2, ncol(geneBack)), function(i) {
lapply(seq(1,i-1), function(j) {
var1 <- geneBack[,i]
var2 <- geneBack[,j]
pval <- tryCatch(chisq.test(var1,var2)$p.value,
error = function(n) NA)
tibble(geneA = colnames(geneBack)[i],
geneB = colnames(geneBack[j]),
p = pval)
}) %>% bind_rows()
}) %>% bind_rows() %>%
mutate(p.adj = p.adjust(p, method = "BH")) %>%
arrange(p)Gene pairs that have significant interactions (5% FDR)
chiTab %>% filter(p.adj < 0.1) %>% print(n=40)# A tibble: 9 × 4
geneA geneB p p.adj
<chr> <chr> <dbl> <dbl>
1 TP53 del17p 0.00000415 0.000956
2 trisomy19 trisomy12 0.00000581 0.000956
3 EGR2 DDX3X 0.00000721 0.000956
4 IgH_break del5IgH 0.0000119 0.00119
5 ATM del11q 0.0000181 0.00144
6 gain8q del8p 0.0000374 0.00248
7 trisomy12 del13q 0.0000439 0.00250
8 FAT4 ATM 0.0000579 0.00288
9 TP53 del5IgH 0.000407 0.0180
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.1 stringr_1.4.1 dplyr_1.0.9
[4] purrr_0.3.4 readr_2.1.2 tidyr_1.2.0
[7] tibble_3.1.8 tidyverse_1.3.2 limma_3.52.2
[10] IHW_1.24.0 readxl_1.4.0 gtable_0.3.0
[13] ggbeeswarm_0.6.0 jyluMisc_0.1.5 colorspace_2.0-3
[16] RColorBrewer_1.1-3 ggrepel_0.9.1 ggplot2_3.3.6
[19] cowplot_1.1.1 genefilter_1.78.0 pheatmap_1.0.12
[22] reshape2_1.4.4 gridExtra_2.3 Biobase_2.56.0
[25] BiocGenerics_0.42.0
loaded via a namespace (and not attached):
[1] utf8_1.2.2 shinydashboard_0.7.2
[3] tidyselect_1.1.2 RSQLite_2.2.15
[5] AnnotationDbi_1.58.0 htmlwidgets_1.5.4
[7] grid_4.2.0 BiocParallel_1.30.3
[9] maxstat_0.7-25 munsell_0.5.0
[11] ragg_1.2.2 codetools_0.2-18
[13] DT_0.23 withr_2.5.0
[15] highr_0.9 knitr_1.39
[17] rstudioapi_0.13 stats4_4.2.0
[19] ggsignif_0.6.3 labeling_0.4.2
[21] MatrixGenerics_1.8.1 git2r_0.30.1
[23] slam_0.1-50 GenomeInfoDbData_1.2.8
[25] lpsymphony_1.24.0 KMsurv_0.1-5
[27] farver_2.1.1 bit64_4.0.5
[29] rprojroot_2.0.3 vctrs_0.4.1
[31] generics_0.1.3 TH.data_1.1-1
[33] xfun_0.31 sets_1.0-21
[35] R6_2.5.1 GenomeInfoDb_1.32.2
[37] bitops_1.0-7 cachem_1.0.6
[39] fgsea_1.22.0 DelayedArray_0.22.0
[41] assertthat_0.2.1 vroom_1.5.7
[43] promises_1.2.0.1 scales_1.2.0
[45] multcomp_1.4-19 googlesheets4_1.0.0
[47] beeswarm_0.4.0 sandwich_3.0-2
[49] workflowr_1.7.0 rlang_1.0.6
[51] systemfonts_1.0.4 splines_4.2.0
[53] rstatix_0.7.0 gargle_1.2.0
[55] broom_1.0.0 BiocManager_1.30.18
[57] yaml_2.3.5 abind_1.4-5
[59] modelr_0.1.8 crosstalk_1.2.0
[61] backports_1.4.1 httpuv_1.6.6
[63] tools_4.2.0 relations_0.6-12
[65] ellipsis_0.3.2 gplots_3.1.3
[67] jquerylib_0.1.4 Rcpp_1.0.9
[69] plyr_1.8.7 visNetwork_2.1.0
[71] zlibbioc_1.42.0 RCurl_1.98-1.7
[73] ggpubr_0.4.0 S4Vectors_0.34.0
[75] zoo_1.8-10 SummarizedExperiment_1.26.1
[77] haven_2.5.0 cluster_2.1.3
[79] exactRankTests_0.8-35 fs_1.5.2
[81] magrittr_2.0.3 data.table_1.14.2
[83] reprex_2.0.1 survminer_0.4.9
[85] googledrive_2.0.0 mvtnorm_1.1-3
[87] matrixStats_0.62.0 hms_1.1.1
[89] shinyjs_2.1.0 mime_0.12
[91] evaluate_0.15 xtable_1.8-4
[93] XML_3.99-0.10 IRanges_2.30.0
[95] compiler_4.2.0 KernSmooth_2.23-20
[97] crayon_1.5.2 htmltools_0.5.3
[99] later_1.3.0 tzdb_0.3.0
[101] lubridate_1.8.0 DBI_1.1.3
[103] dbplyr_2.2.1 MASS_7.3-58
[105] BiocStyle_2.24.0 Matrix_1.4-1
[107] car_3.1-0 cli_3.4.1
[109] marray_1.74.0 parallel_4.2.0
[111] igraph_1.3.4 GenomicRanges_1.48.0
[113] pkgconfig_2.0.3 km.ci_0.5-6
[115] piano_2.12.0 xml2_1.3.3
[117] annotate_1.74.0 vipor_0.4.5
[119] bslib_0.4.1 XVector_0.36.0
[121] drc_3.0-1 rvest_1.0.2
[123] digest_0.6.30 Biostrings_2.64.0
[125] rmarkdown_2.14 cellranger_1.1.0
[127] fastmatch_1.1-3 survMisc_0.5.6
[129] shiny_1.7.3 gtools_3.9.3
[131] lifecycle_1.0.3 jsonlite_1.8.3
[133] carData_3.0-5 fansi_1.0.3
[135] pillar_1.8.0 lattice_0.20-45
[137] KEGGREST_1.36.3 fastmap_1.1.0
[139] httr_1.4.3 plotrix_3.8-2
[141] survival_3.4-0 glue_1.6.2
[143] fdrtool_1.2.17 png_0.1-7
[145] bit_4.0.4 stringi_1.7.8
[147] sass_0.4.2 blob_1.2.3
[149] textshaping_0.3.6 caTools_1.18.2
[151] memoise_2.0.1