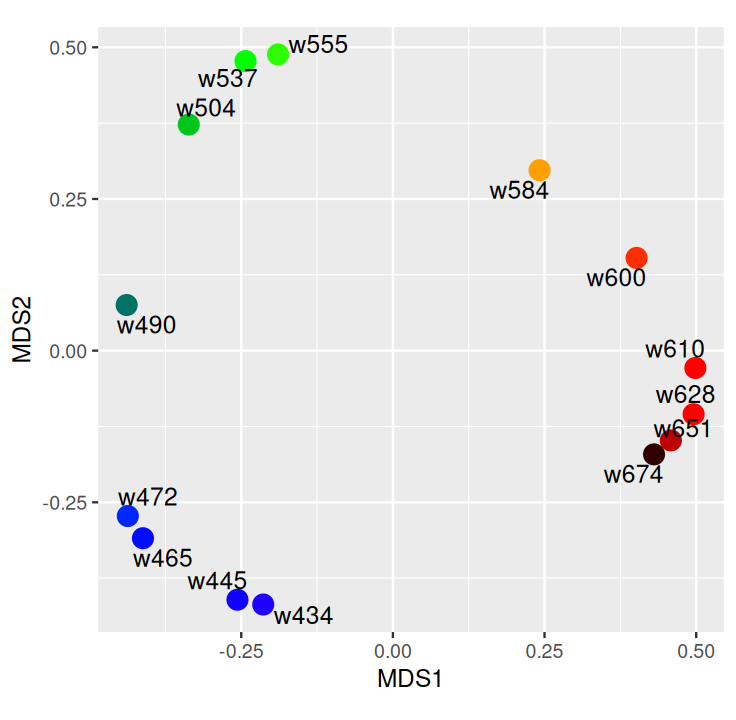
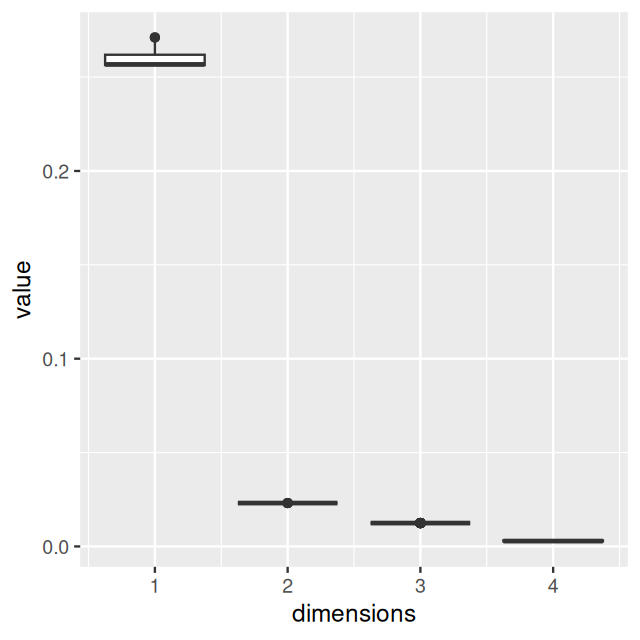
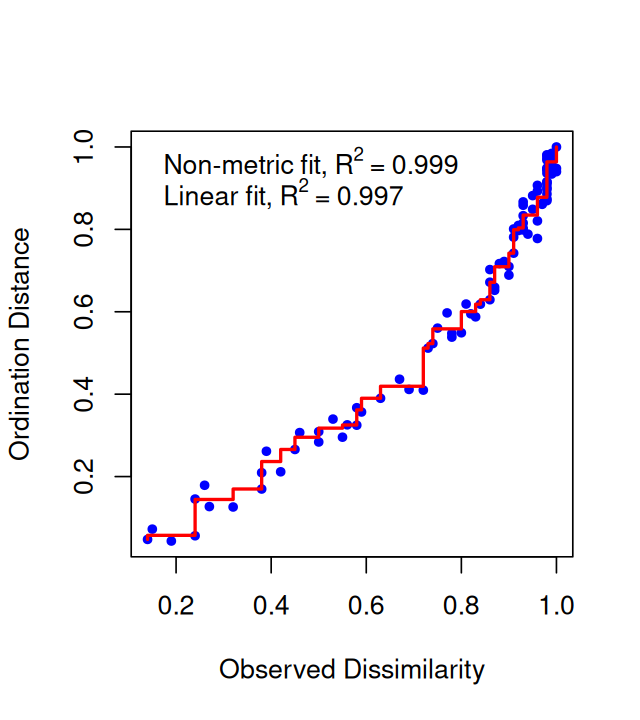
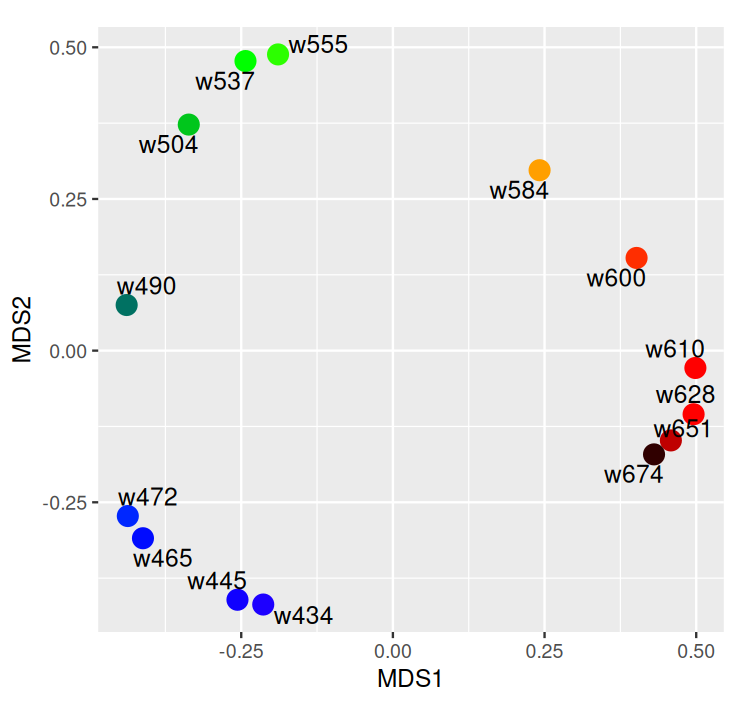
Belkin, Mikhail, and Partha Niyogi. 2003. “Laplacian Eigenmaps for Dimensionality Reduction and Data Representation.” Neural Computation 15 (6): 1373–96.
Bengio, Yoshua, Jean-François Paiement, Pascal Vincent, Olivier Delalleau, Nicolas Le Roux, and Marie Ouimet. 2004. “Out-of-Sample Extensions for LLE, Isomap, MDS, Eigenmaps, and Spectral Clustering.” Advances in Neural Information Processing Systems 16: 177–84.
Braak, Cajo ter. 1985. “Correspondence Analysis of Incidence and Abundance Data: Properties in Terms of a Unimodal Respose.” Biometrics 41 (January).
Brodie, Eoin L, Todd Z DeSantis, Dominique C Joyner, Seung M Baek, Joern T Larsen, Gary L Andersen, Terry C Hazen, et al. 2006. “Application of a High-Density Oligonucleotide Microarray Approach to Study Bacterial Population Dynamics During Uranium Reduction and Reoxidation.” Applied and Environmental Microbiology 72 (9): 6288–98.
Callahan, Ben J, Kris Sankaran, Julia A Fukuyama, Paul J McMurdie, and Susan P Holmes. 2016. “Bioconductor Workflow for Microbiome Data Analysis: From Raw Reads to Community Analyses.” F1000Research 5.
Chessel, Daniel, Anne Dufour, and Jean Thioulouse. 2004.
“The ade4 Package - i: One-Table Methods.” R News 4 (1): 5–10.
http://CRAN.R-project.org/doc/Rnews/.
Diaconis, Persi, Sharad Goel, and Susan Holmes. 2008.
“Horseshoes in Multidimensional Scaling and Kernel Methods.” Annals of Applied Statistics 2: 777.
https://doi.org/DOI:10.1214/08-AOAS165.
Ekman, Gosta. 1954. “Dimensions of Color Vision.” The Journal of Psychology 38 (2): 467–74.
Goslee, Sarah C, Dean L Urban, et al. 2007. “The Ecodist Package for Dissimilarity-Based Analysis of Ecological Data.” Journal of Statistical Software 22 (7): 1–19.
Greenacre, Michael J. 2007. Correspondence Analysis in Practice. Chapman & Hall.
Guillot, Gilles, and François Rousset. 2013. “Dismantling the Mantel Tests.” Methods in Ecology and Evolution 4 (4): 336–44.
Hastie, Trevor, and Werner Stuetzle. 1989. “Principal Curves.” Journal of the American Statistical Association 84 (406): 502–16.
Hastie, Trevor, Robert Tibshirani, and Jerome Friedman. 2008. The Elements of Statistical Learning. 2^{\text{nd}} ed. Springer.
Holmes, Susan. 2006.
“Multivariate Analysis: The French way.” In
Probability and Statistics: Essays in Honor of David a. Freedman, edited by D. Nolan and T. P. Speed. Vol. 56. IMS Lecture Notes–Monograph Series. Beachwood, OH: IMS.
http://www.imstat.org/publications/lecnotes.htm.
Holmes, Susan, Alexander V Alekseyenko, Alden Timme, Tyrrell Nelson, Pankaj Jay Pasricha, and Alfred Spormann. 2011. “Visualization and Statistical Comparisons of Microbial Communities Using r Packages on Phylochip Data.” In Pacific Symposium on Biocomputing, 142–53. World Scientific.
Izenman, Alan Julian. 2008. “Nonlinear Dimensionality Reduction and Manifold Learning.” In Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning, 597–632. New York, NY: Springer New York.
Josse, Julie, and Susan Holmes. 2016. “Measuring Multivariate Association and Beyond.” Statistics Surveys 10: 132–67.
Kashyap, Purna C, Angela Marcobal, Luke K Ursell, Samuel A Smits, Erica D Sonnenburg, Elizabeth K Costello, Steven K Higginbottom, et al. 2013. “Genetically Dictated Change in Host Mucus Carbohydrate Landscape Exerts a Diet-Dependent Effect on the Gut Microbiota.” PNAS 110 (42): 17059–64.
Kendall, David. 1969. “Incidence Matrices, Interval Graphs and Seriation in Archeology.” Pacific Journal of Mathematics 28 (3): 565–70.
Leek, Jeffrey T, Robert B Scharpf, Héctor Corrada Bravo, David Simcha, Benjamin Langmead, W Evan Johnson, Donald Geman, Keith Baggerly, and Rafael A Irizarry. 2010. “Tackling the Widespread and Critical Impact of Batch Effects in High-Throughput Data.” Nature Reviews Genetics 11 (10): 733–39.
Moignard, Victoria, Steven Woodhouse, Laleh Haghverdi, Andrew J Lilly, Yosuke Tanaka, Adam C Wilkinson, Florian Buettner, et al. 2015. “Decoding the Regulatory Network of Early Blood Development from Single-Cell Gene Expression Measurements.” Nature Biotechnology.
Nelson, Tyrell A, Susan Holmes, Alexander Alekseyenko, Masha Shenoy, Todd DeSantis, Cindy Wu, Gary Andersen, et al. 2010. “PhyloChip Microarray Analysis Reveals Altered Gastrointestinal Microbial Communities in a Rat Model of Colonic Hypersensitivity.” Neurogastroenterology & Motility.
Pagès, Jérôme. 2016. Multiple Factor Analysis by Example Using R. CRC Press.
Perraudeau, Fanny, Davide Risso, Kelly Street, Elizabeth Purdom, and Sandrine Dudoit. 2017. “Bioconductor Workflow for Single-Cell RNA Sequencing: Normalization, Dimensionality Reduction, Clustering, and Lineage Inference.” F1000Research 6.
Prentice, IC. 1977. “Non-Metric Ordination Methods in Ecology.” The Journal of Ecology, 85–94.
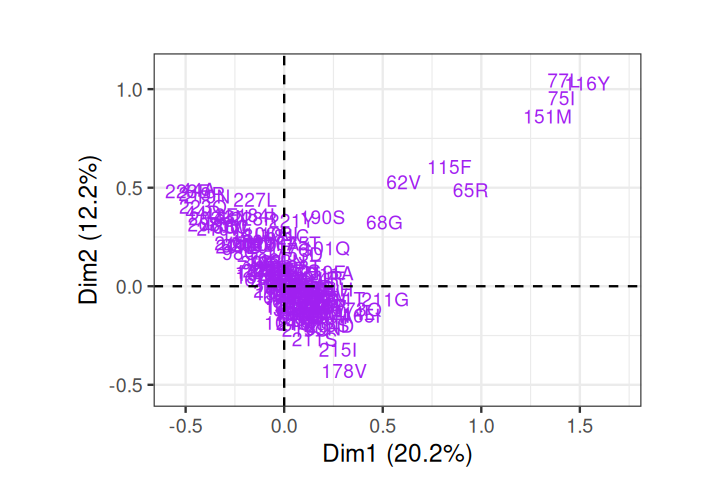
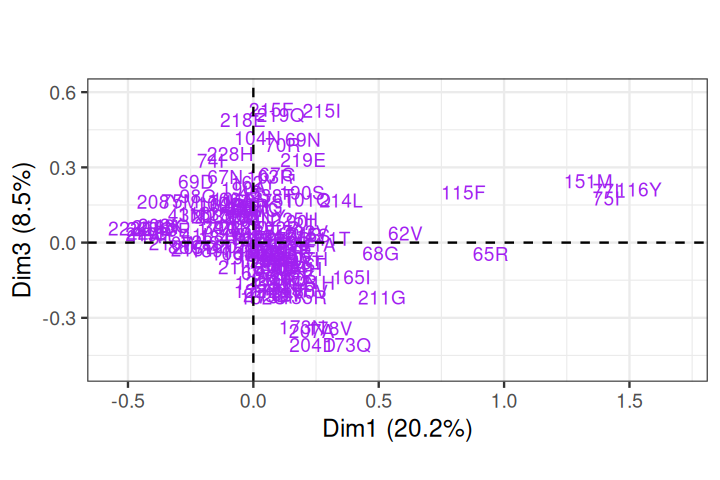
Rhee, Soo-Yon, Matthew J Gonzales, Rami Kantor, Bradley J Betts, Jaideep Ravela, and Robert W Shafer. 2003. “Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database.” Nucleic Acids Research 31 (1): 298–303.
Roweis, Sam T, and Lawrence K Saul. 2000. “Nonlinear Dimensionality Reduction by Locally Linear Embedding.” Science 290 (5500): 2323–26.
Tenenbaum, Joshua B, Vin De Silva, and John C Langford. 2000. “A Global Geometric Framework for Nonlinear Dimensionality Reduction.” Science 290 (5500): 2319–23.
Trosset, Michael W, and Carey E Priebe. 2008. “The Out-of-Sample Problem for Classical Multidimensional Scaling.” Computational Statistics & Data Analysis 52 (10): 4635–42.